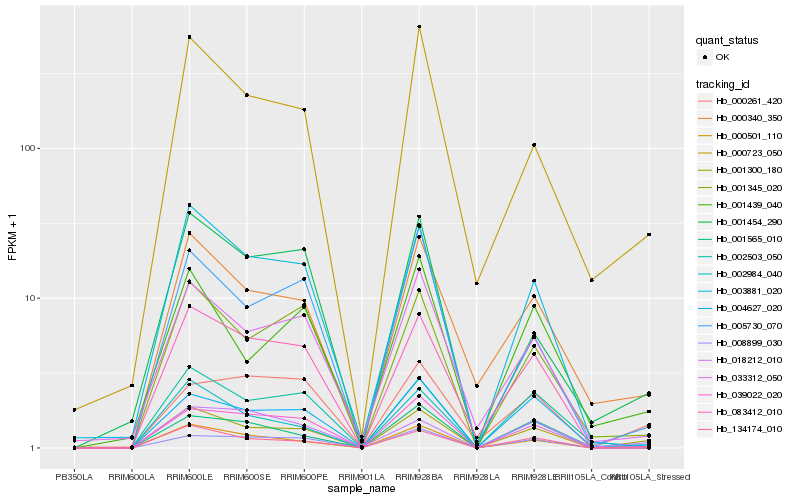
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001300_180 |
0.0 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 2 |
Hb_134174_010 |
0.0938521616 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_008899_030 |
0.1290163155 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 4 |
Hb_000340_350 |
0.1453206481 |
- |
- |
PREDICTED: sugar transporter ERD6-like 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004627_020 |
0.1540852925 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 6 |
Hb_001345_020 |
0.1558681686 |
- |
- |
hypothetical protein POPTR_0007s10400g [Populus trichocarpa] |
| 7 |
Hb_000723_050 |
0.1602538031 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 8 |
Hb_002503_050 |
0.1626527809 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 9 |
Hb_003881_020 |
0.1638626296 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 10 |
Hb_083412_010 |
0.1656030693 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 11 |
Hb_033312_050 |
0.1658306242 |
- |
- |
PREDICTED: uncharacterized protein LOC105638598, partial [Jatropha curcas] |
| 12 |
Hb_000501_110 |
0.1676051004 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 [Populus euphratica] |
| 13 |
Hb_002984_040 |
0.1682796306 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_001439_040 |
0.169260039 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001565_010 |
0.1725703834 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 16 |
Hb_001454_290 |
0.1775166122 |
- |
- |
PREDICTED: uncharacterized protein LOC105643442 [Jatropha curcas] |
| 17 |
Hb_018212_010 |
0.1786488425 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 18 |
Hb_005730_070 |
0.1795858642 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 8 [Jatropha curcas] |
| 19 |
Hb_039022_020 |
0.1805331273 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 20 |
Hb_000261_420 |
0.1807163952 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |