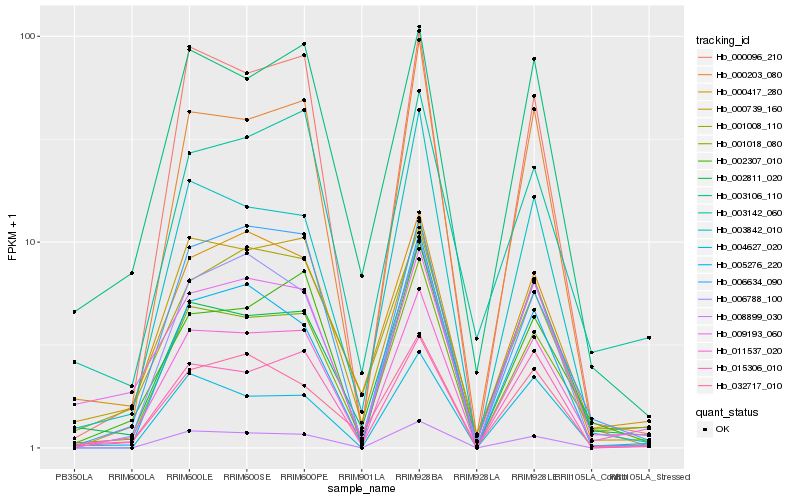
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011537_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 2 |
Hb_001018_080 |
0.0595084283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008899_030 |
0.1011464653 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 4 |
Hb_000739_160 |
0.1146783468 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002307_010 |
0.1149021672 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 6 |
Hb_004627_020 |
0.1165984006 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 7 |
Hb_015306_010 |
0.1170074876 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 8 |
Hb_000203_080 |
0.1181907909 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 9 |
Hb_002811_020 |
0.1194046974 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 10 |
Hb_006788_100 |
0.1230852709 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000096_210 |
0.1239836201 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 12 |
Hb_003142_060 |
0.1281966889 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_009193_060 |
0.1284230629 |
- |
- |
PREDICTED: ureide permease 1-like [Jatropha curcas] |
| 14 |
Hb_000417_280 |
0.1287085041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_032717_010 |
0.1299840358 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_006634_090 |
0.1301666127 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 17 |
Hb_005276_220 |
0.1310055721 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 18 |
Hb_001008_110 |
0.1313403163 |
- |
- |
- |
| 19 |
Hb_003842_010 |
0.1315869052 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_003106_110 |
0.132787011 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |