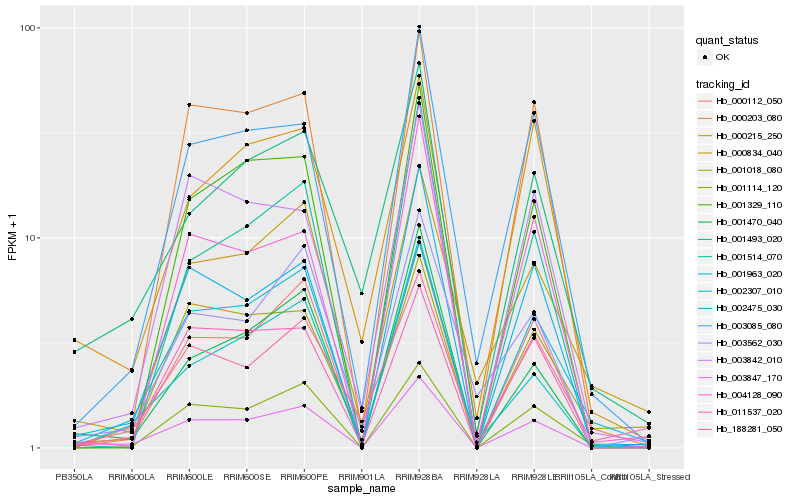
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003847_170 |
0.0 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 2 |
Hb_000112_050 |
0.1227918167 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 3 |
Hb_001470_040 |
0.1233517241 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 4 |
Hb_002307_010 |
0.1248010593 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 5 |
Hb_001963_020 |
0.1326835349 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002475_030 |
0.1340485347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001114_120 |
0.1439355558 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_011537_020 |
0.1481854338 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 9 |
Hb_003085_080 |
0.1484533391 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_001329_110 |
0.1501485963 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 11 |
Hb_001018_080 |
0.1506906989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000203_080 |
0.1513121848 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 13 |
Hb_188281_050 |
0.1522160897 |
- |
- |
PREDICTED: solute carrier family 35 member F1-like isoform X1 [Vitis vinifera] |
| 14 |
Hb_001493_020 |
0.1522493627 |
- |
- |
leucine-rich repeat receptor-like protein kinase [Manihot esculenta] |
| 15 |
Hb_003562_030 |
0.152869732 |
- |
- |
- |
| 16 |
Hb_003842_010 |
0.1561163334 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 17 |
Hb_004128_090 |
0.1592431538 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 18 |
Hb_001514_070 |
0.1600917024 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000215_250 |
0.1606189941 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000834_040 |
0.1610715243 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |