| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
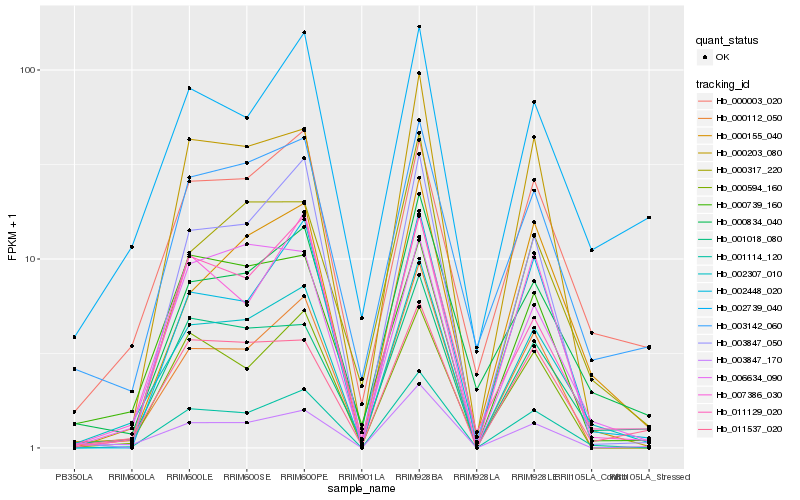
Hb_002307_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 2 |
Hb_001114_120 |
0.0816322324 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_001018_080 |
0.1111410722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011537_020 |
0.1149021672 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 5 |
Hb_000112_050 |
0.1166250376 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 6 |
Hb_000834_040 |
0.1240661501 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 7 |
Hb_003847_170 |
0.1248010593 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 8 |
Hb_000003_020 |
0.1276351567 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 9 |
Hb_000203_080 |
0.128337941 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 10 |
Hb_003142_060 |
0.1288266316 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_003847_050 |
0.1310410033 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 12 |
Hb_000155_040 |
0.1313255348 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 13 |
Hb_000317_220 |
0.1333587941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006634_090 |
0.1351369573 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 15 |
Hb_007386_030 |
0.1357835536 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 16 |
Hb_000739_160 |
0.1367570577 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_011129_020 |
0.1381655544 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 18 |
Hb_002739_040 |
0.1381868838 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 19 |
Hb_002448_020 |
0.1425386372 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000594_160 |
0.143649405 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |