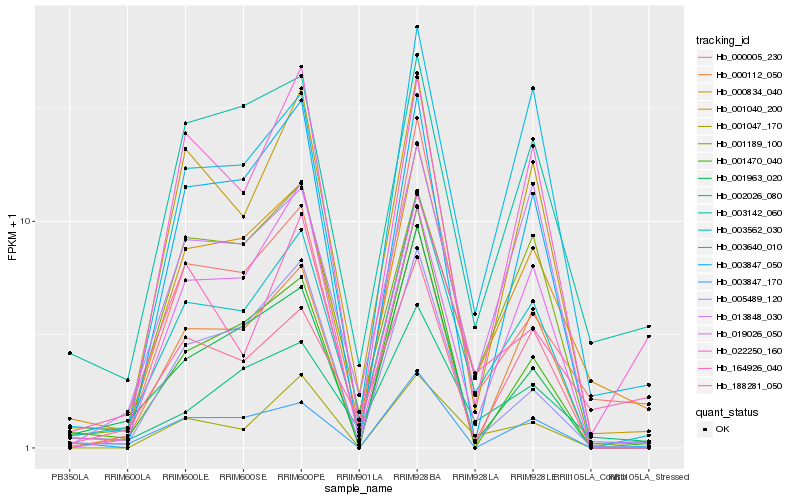
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003562_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000834_040 |
0.1118629423 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 3 |
Hb_001040_200 |
0.1268093504 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 4 |
Hb_188281_050 |
0.1325482981 |
- |
- |
PREDICTED: solute carrier family 35 member F1-like isoform X1 [Vitis vinifera] |
| 5 |
Hb_000112_050 |
0.1360914536 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 6 |
Hb_003640_010 |
0.1440474812 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 7 |
Hb_001047_170 |
0.1449593231 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 8 |
Hb_001189_100 |
0.1456316889 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 9 |
Hb_013848_030 |
0.1456497205 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 10 |
Hb_001963_020 |
0.1468744308 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003847_170 |
0.152869732 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 12 |
Hb_003142_060 |
0.1568466584 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_003847_050 |
0.158155537 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 14 |
Hb_005489_120 |
0.15837739 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 15 |
Hb_019026_050 |
0.1590229899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002026_080 |
0.1592054556 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 17 |
Hb_164926_040 |
0.1624038027 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 18 |
Hb_000005_230 |
0.16266852 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 19 |
Hb_001470_040 |
0.1633233964 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 20 |
Hb_022250_160 |
0.1634002084 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |