Hb_002301_110
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig2301: 123306-124070 |
| Sequence |   |
Annotation
kegg
| ID | pop:POPTR_0011s05640g |
|---|---|
| description | hypothetical protein |
nr
| ID | XP_011001231.1 |
|---|---|
| description | PREDICTED: protein E6-like [Populus euphratica] |
swissprot
| ID | Q01197 |
|---|---|
| description | Protein E6 OS=Gossypium hirsutum GN=E6 PE=2 SV=1 |
trembl
| ID | A9PDV3 |
|---|---|
| description | Uncharacterized protein OS=Populus trichocarpa GN=POPTR_0011s05640g PE=2 SV=1 |
Gene Ontology
| ID | - |
|---|---|
| description | - |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_24124: 123265-124319 |
|---|---|
| cDNA (Sanger) (ID:Location) |
002_D19.ab1: 123265-124070 , 004_J03.ab1: 123265-124063 , 006_P22.ab1: 123265-123991 , 010_B11.ab1: 123265-123893 , 010_K22.ab1: 123265-123753 , 012_M15.ab1: 123265-124008 , 013_E04.ab1: 123265-123917 , 013_J13.ab1: 123265-124010 , 015_A14.ab1: 123265-124034 , 015_M18.ab1: 123287-123939 , 016_G12.ab1: 123263-123992 , 021_K01.ab1: 123265-124019 , 028_P22.ab1: 123265-123980 , 030_N04.ab1: 123265-124080 , 037_L16.ab1: 123263-124039 , 040_B13.ab1: 123265-124007 , 042_E08.ab1: 123265-124018 , 042_N10.ab1: 123265-124046 , 044_D13.ab1: 123265-124072 , 046_A22.ab1: 123265-123820 , 051_H23.ab1: 123265-124042 , 052_E03.ab1: 123265-124011 |
Similar expressed genes (Top20)
Gene co-expression network
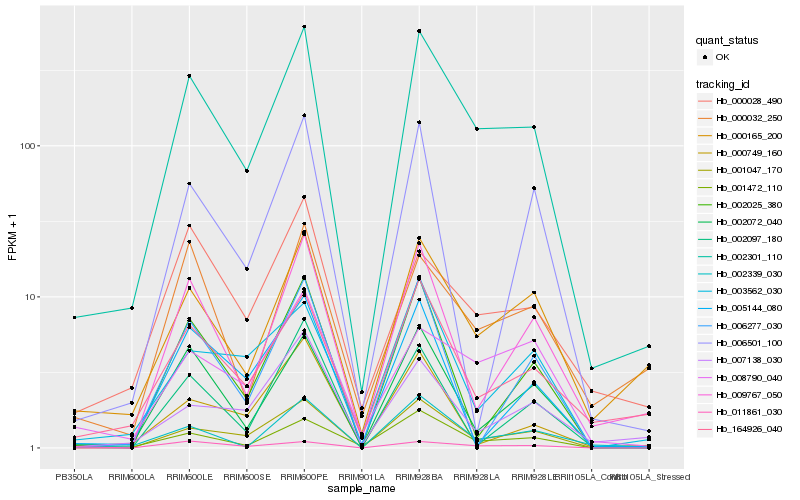
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 7.43458 | 67.2743 | 289.809 | 614.733 | 6.30259 | 1.34399 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 2.3643 | 3.71785 | 128.657 | 574.429 | 132.14 |

