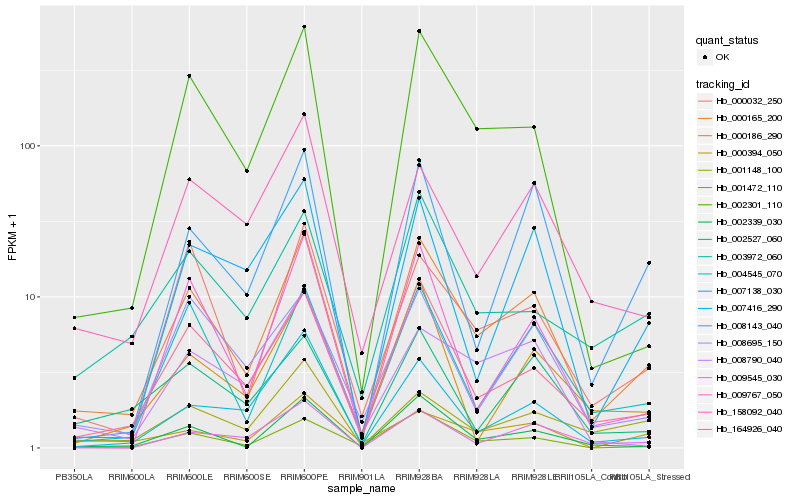
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_200 |
0.0 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_164926_040 |
0.1268739241 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 3 |
Hb_001472_110 |
0.1361290146 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 4 |
Hb_004545_070 |
0.1447159065 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_002301_110 |
0.1475612418 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 6 |
Hb_002339_030 |
0.1525313556 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_009545_030 |
0.1571212714 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000032_250 |
0.1631516485 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 9 |
Hb_008143_040 |
0.1708606633 |
- |
- |
Alpha-expansin 4 precursor, putative [Ricinus communis] |
| 10 |
Hb_158092_040 |
0.1710196775 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 11 |
Hb_003972_060 |
0.1711516272 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 12 |
Hb_001148_100 |
0.1719629026 |
- |
- |
- |
| 13 |
Hb_007138_030 |
0.1734392904 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 14 |
Hb_009767_050 |
0.1739220694 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008695_150 |
0.1751524088 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000394_050 |
0.1810675514 |
- |
- |
- |
| 17 |
Hb_007416_290 |
0.1813280982 |
- |
- |
PREDICTED: plasma membrane ATPase 4-like [Pyrus x bretschneideri] |
| 18 |
Hb_008790_040 |
0.1819583567 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Jatropha curcas] |
| 19 |
Hb_002527_060 |
0.182681043 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 20 |
Hb_000186_290 |
0.1850253766 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |