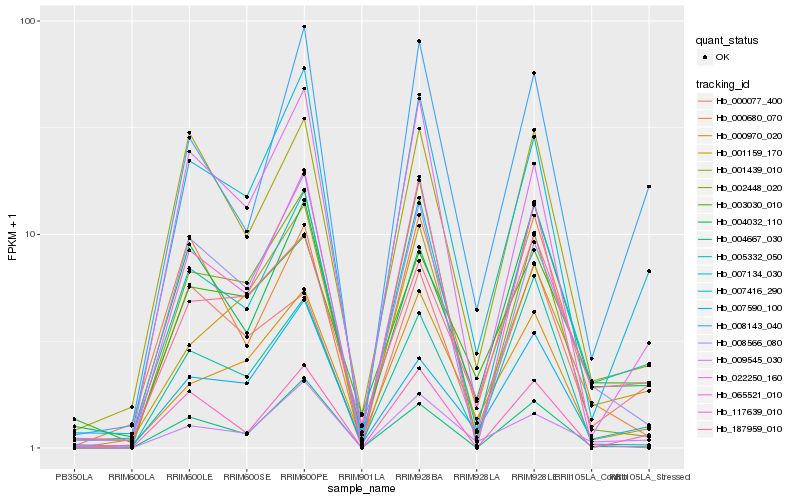
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007134_030 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 2 |
Hb_007416_290 |
0.1256056733 |
- |
- |
PREDICTED: plasma membrane ATPase 4-like [Pyrus x bretschneideri] |
| 3 |
Hb_008143_040 |
0.1256556726 |
- |
- |
Alpha-expansin 4 precursor, putative [Ricinus communis] |
| 4 |
Hb_187959_010 |
0.1273468526 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 5 |
Hb_004032_110 |
0.1304198741 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 6 |
Hb_009545_030 |
0.1378688164 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001439_010 |
0.139525335 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 8 |
Hb_000970_020 |
0.1406346681 |
- |
- |
hypothetical protein POPTR_0005s19050g [Populus trichocarpa] |
| 9 |
Hb_008566_080 |
0.1437552764 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 10 |
Hb_001159_170 |
0.1445579878 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 11 |
Hb_007590_100 |
0.1471129288 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 12 |
Hb_000077_400 |
0.1490228207 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_117639_010 |
0.1492885743 |
- |
- |
hypothetical protein B456_013G070100 [Gossypium raimondii] |
| 14 |
Hb_000680_070 |
0.1531681266 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004667_030 |
0.1532837225 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 16 |
Hb_002448_020 |
0.1544879003 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 17 |
Hb_022250_160 |
0.1555894448 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 18 |
Hb_003030_010 |
0.1561639092 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 19 |
Hb_005332_050 |
0.1601352574 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 20 |
Hb_065521_010 |
0.1629230462 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |