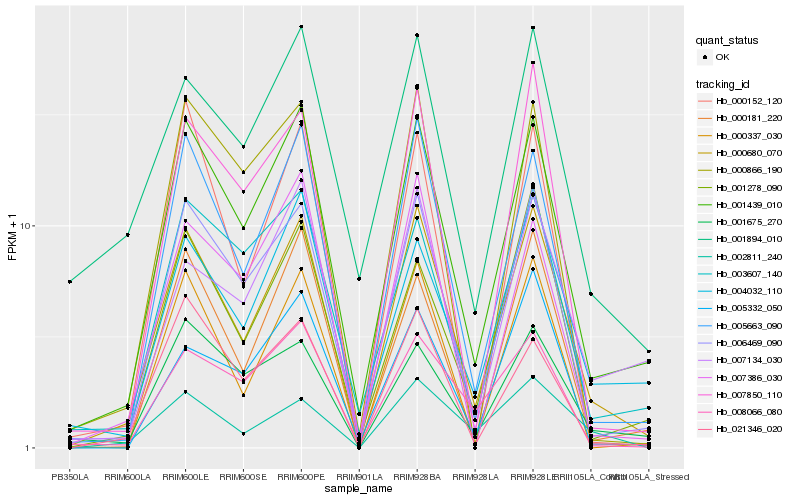
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000680_070 |
0.0 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001439_010 |
0.0831478638 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 3 |
Hb_005663_090 |
0.099725573 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 4 |
Hb_006469_090 |
0.1021622416 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 5 |
Hb_000337_030 |
0.1231845241 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_007850_110 |
0.1327613358 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 7 |
Hb_004032_110 |
0.1400404236 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002811_240 |
0.1414670572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000181_220 |
0.143849557 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001278_090 |
0.1456555828 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 11 |
Hb_008066_080 |
0.1487059411 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 12 |
Hb_001894_010 |
0.1497150355 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 13 |
Hb_000866_190 |
0.1503677884 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000152_120 |
0.1522745566 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 15 |
Hb_021346_020 |
0.1529656883 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 16 |
Hb_007134_030 |
0.1531681266 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 17 |
Hb_007386_030 |
0.153602275 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 18 |
Hb_001675_270 |
0.1536836777 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 19 |
Hb_003607_140 |
0.1542973887 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 20 |
Hb_005332_050 |
0.1561038643 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |