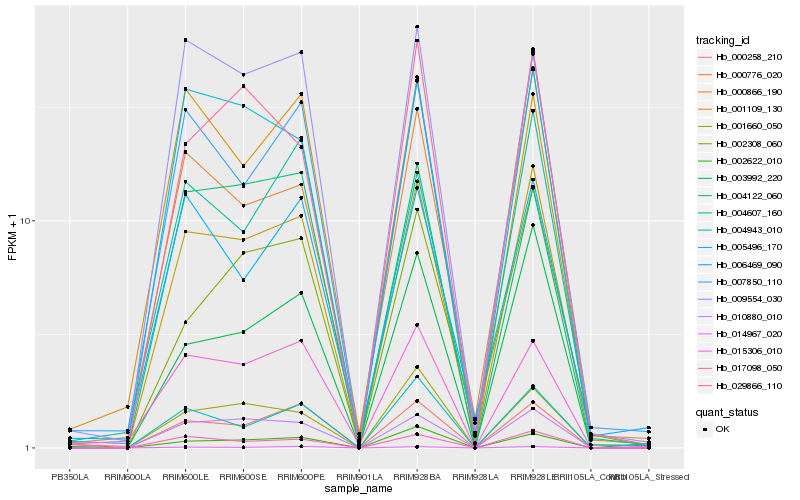
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001109_130 |
0.0 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 136 isoform X1 [Jatropha curcas] |
| 2 |
Hb_017098_050 |
0.0769279043 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 3 |
Hb_010880_010 |
0.0813065082 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000258_210 |
0.0882534322 |
- |
- |
prenyl-dependent CAAX protease, putative [Ricinus communis] |
| 5 |
Hb_007850_110 |
0.0892209464 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 6 |
Hb_004943_010 |
0.1076872625 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_005496_170 |
0.1080271825 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 8 |
Hb_014967_020 |
0.111405743 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_000776_020 |
0.1132684309 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 10 |
Hb_003992_220 |
0.1152153402 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 11 |
Hb_002622_010 |
0.1162452988 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 12 |
Hb_009554_030 |
0.1213769656 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 13 |
Hb_001660_050 |
0.1235297204 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Eucalyptus grandis] |
| 14 |
Hb_015306_010 |
0.1241976464 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 15 |
Hb_029866_110 |
0.1256255404 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 16 |
Hb_004607_160 |
0.1261656617 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 17 |
Hb_002308_060 |
0.1280500467 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 18 |
Hb_000866_190 |
0.1310743758 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_004122_060 |
0.1331360775 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 20 |
Hb_006469_090 |
0.1332500183 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |