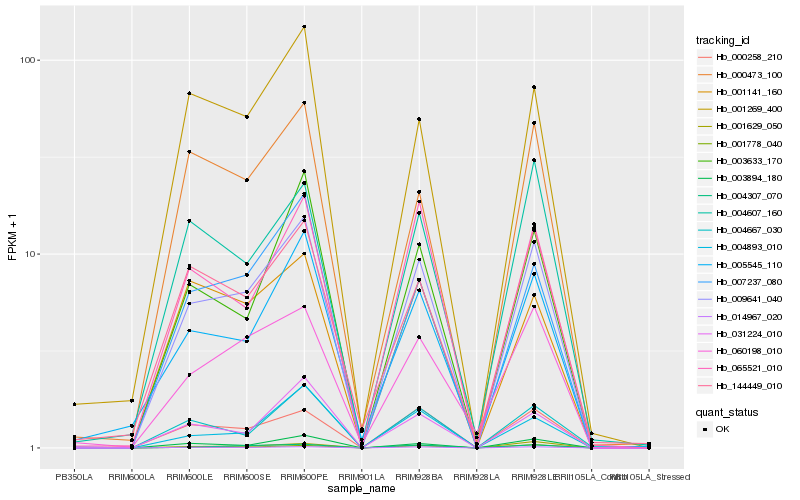
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009641_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 2 |
Hb_014967_020 |
0.0779308819 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_000258_210 |
0.1047435757 |
- |
- |
prenyl-dependent CAAX protease, putative [Ricinus communis] |
| 4 |
Hb_007237_080 |
0.1119250284 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_003894_180 |
0.1134469278 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 6 |
Hb_004307_070 |
0.116379028 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_003633_170 |
0.1178913436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005545_110 |
0.1203843731 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 9 |
Hb_065521_010 |
0.1208839837 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 10 |
Hb_060198_010 |
0.1282686462 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 11 |
Hb_000473_100 |
0.1313071209 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_031224_010 |
0.13188731 |
- |
- |
- |
| 13 |
Hb_004667_030 |
0.1321959968 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 14 |
Hb_144449_010 |
0.132294357 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 15 |
Hb_001269_400 |
0.132401194 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 16 |
Hb_001629_050 |
0.1333477528 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 17 |
Hb_004893_010 |
0.1348397232 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 18 |
Hb_001141_160 |
0.1368157042 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 19 |
Hb_004607_160 |
0.1368514806 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 20 |
Hb_001778_040 |
0.1383976555 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |