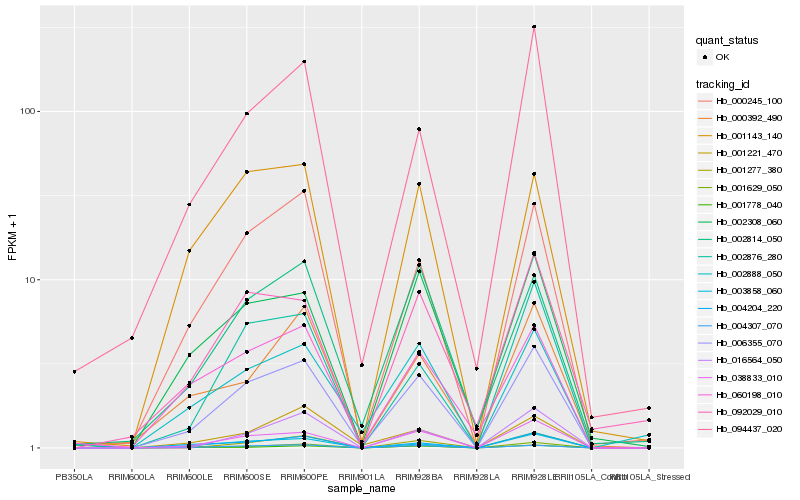
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006355_070 |
0.0 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 2 |
Hb_038833_010 |
0.0843327847 |
- |
- |
polyprotein [Ananas comosus] |
| 3 |
Hb_004307_070 |
0.1058075933 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000245_100 |
0.1197988101 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 5 |
Hb_003858_060 |
0.1277718127 |
- |
- |
Germin-like protein subfamily 1 member 18 [Theobroma cacao] |
| 6 |
Hb_002876_280 |
0.1356735405 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 7 |
Hb_001629_050 |
0.1376532256 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 8 |
Hb_004204_220 |
0.140178462 |
- |
- |
- |
| 9 |
Hb_002308_060 |
0.1410558185 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 10 |
Hb_002888_050 |
0.1410627892 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001277_380 |
0.1439023437 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 3 [Tarenaya hassleriana] |
| 12 |
Hb_002814_050 |
0.144878484 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 13 |
Hb_094437_020 |
0.1457476759 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_001221_470 |
0.1499315613 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 15 |
Hb_060198_010 |
0.1500105988 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 16 |
Hb_001143_140 |
0.1511928995 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001778_040 |
0.153672089 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 18 |
Hb_000392_490 |
0.1575688846 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 19 |
Hb_016564_050 |
0.1592673232 |
- |
- |
- |
| 20 |
Hb_092029_010 |
0.1632794497 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |