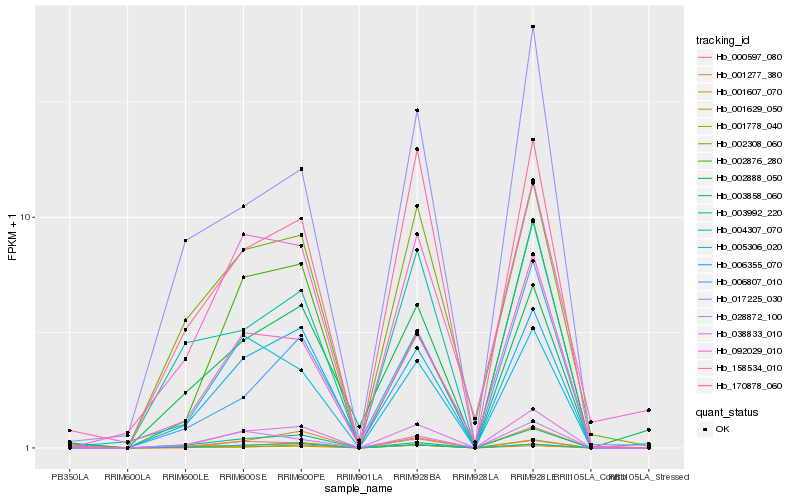
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038833_010 |
0.0 |
- |
- |
polyprotein [Ananas comosus] |
| 2 |
Hb_006355_070 |
0.0843327847 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 3 |
Hb_170878_060 |
0.1217893923 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 4 |
Hb_002308_060 |
0.1332526425 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 5 |
Hb_028872_100 |
0.1440545268 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 6 |
Hb_158534_010 |
0.1458086568 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 7 |
Hb_001277_380 |
0.1470360765 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 3 [Tarenaya hassleriana] |
| 8 |
Hb_003992_220 |
0.1490292591 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 9 |
Hb_092029_010 |
0.1497500146 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 10 |
Hb_002876_280 |
0.1506578279 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 11 |
Hb_006807_010 |
0.1514292209 |
- |
- |
hypothetical protein JCGZ_20881 [Jatropha curcas] |
| 12 |
Hb_000597_080 |
0.1530506529 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_004307_070 |
0.1544968372 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_003858_060 |
0.1573437227 |
- |
- |
Germin-like protein subfamily 1 member 18 [Theobroma cacao] |
| 15 |
Hb_001778_040 |
0.1580589624 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 16 |
Hb_001607_070 |
0.1585087793 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 17 |
Hb_001629_050 |
0.1591528078 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 18 |
Hb_005306_020 |
0.1656529139 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 19 |
Hb_002888_050 |
0.1695584147 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_017225_030 |
0.1697257197 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |