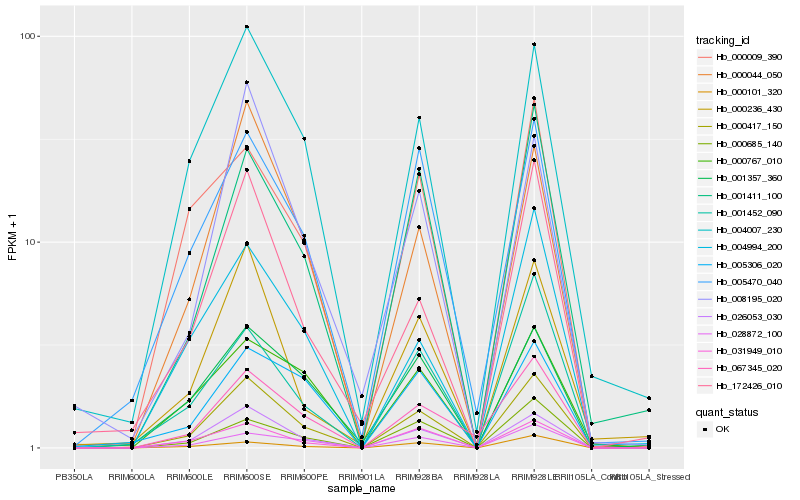
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 2 |
Hb_026053_030 |
0.0679965926 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 3 |
Hb_028872_100 |
0.1111233654 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 4 |
Hb_004007_230 |
0.1174745909 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 5 |
Hb_031949_010 |
0.1193311382 |
- |
- |
- |
| 6 |
Hb_000044_050 |
0.1224682292 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001411_100 |
0.1287605299 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_000685_140 |
0.1298860921 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 9 |
Hb_067345_020 |
0.1336385331 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 10 |
Hb_172426_010 |
0.1359186084 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 11 |
Hb_004994_200 |
0.1381897155 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 12 |
Hb_005470_040 |
0.1385192059 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 13 |
Hb_001452_090 |
0.1393574637 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 14 |
Hb_000236_430 |
0.1425852017 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 15 |
Hb_001357_360 |
0.1428230738 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000101_320 |
0.1454382277 |
- |
- |
- |
| 17 |
Hb_005306_020 |
0.1474835665 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 18 |
Hb_008195_020 |
0.1489962216 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 19 |
Hb_000767_010 |
0.1497818201 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 20 |
Hb_000009_390 |
0.1500427234 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |