| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
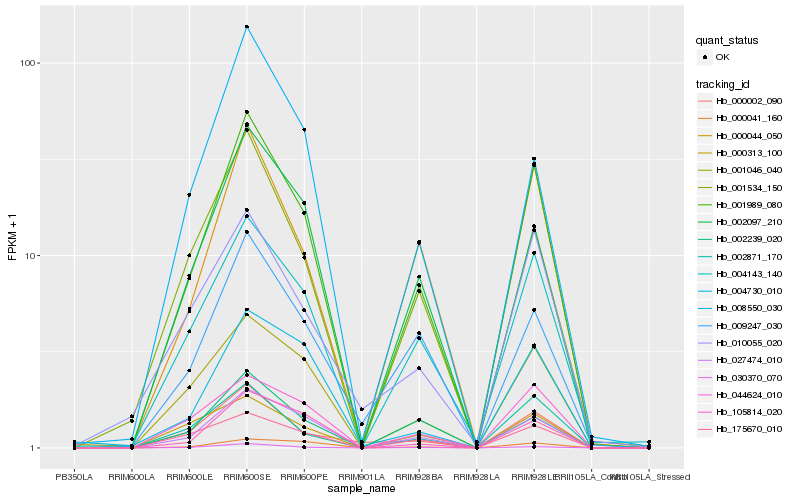
Hb_001989_080 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_027474_010 |
0.0887133456 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 3 |
Hb_004143_140 |
0.0901605203 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 4 |
Hb_000044_050 |
0.1106991514 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001046_040 |
0.1189714074 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_030370_070 |
0.1207281142 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_044624_010 |
0.1249725413 |
- |
- |
- |
| 8 |
Hb_175670_010 |
0.1302402219 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_002871_170 |
0.1303409803 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 10 |
Hb_002097_210 |
0.1318182494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001534_150 |
0.1382830359 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 12 |
Hb_105814_020 |
0.1493535139 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 13 |
Hb_000313_100 |
0.1499137677 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 14 |
Hb_009247_030 |
0.1502666336 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 15 |
Hb_002239_020 |
0.1556622721 |
- |
- |
MLO, putative [Theobroma cacao] |
| 16 |
Hb_000041_160 |
0.1561903443 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_000002_090 |
0.1565264827 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_004730_010 |
0.157052508 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 19 |
Hb_010055_020 |
0.1583889692 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 20 |
Hb_008550_030 |
0.1589210636 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |