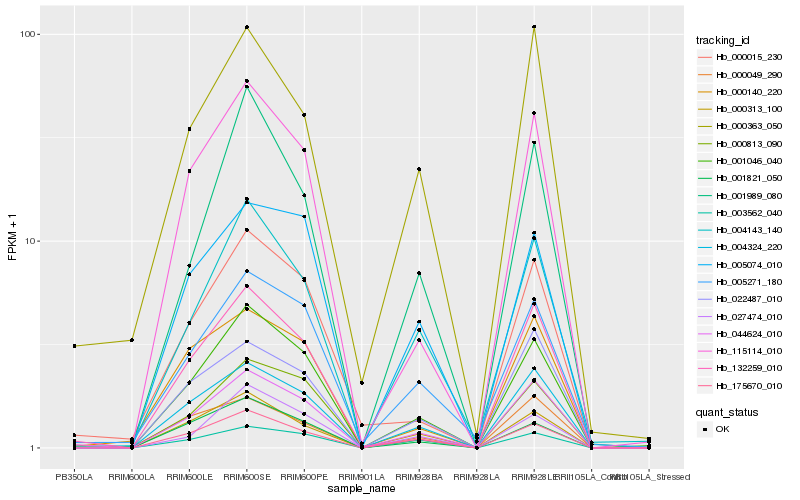
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044624_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004324_220 |
0.0470377055 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 3 |
Hb_001046_040 |
0.0652958812 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_022487_010 |
0.0797822677 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 5 |
Hb_175670_010 |
0.0908784217 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_115114_010 |
0.0943076893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000813_090 |
0.0992226151 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 8 |
Hb_004143_140 |
0.1022706615 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 9 |
Hb_005074_010 |
0.1129831946 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 10 |
Hb_000313_100 |
0.1136092096 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 11 |
Hb_005271_180 |
0.1176570461 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 12 |
Hb_003562_040 |
0.1199607686 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 13 |
Hb_000049_290 |
0.121639996 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_000140_220 |
0.1220517574 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 15 |
Hb_001989_080 |
0.1249725413 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_132259_010 |
0.126988119 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 17 |
Hb_000363_050 |
0.1274759044 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_027474_010 |
0.1295427429 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 19 |
Hb_001821_050 |
0.1368799628 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 20 |
Hb_000015_230 |
0.1376810621 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |