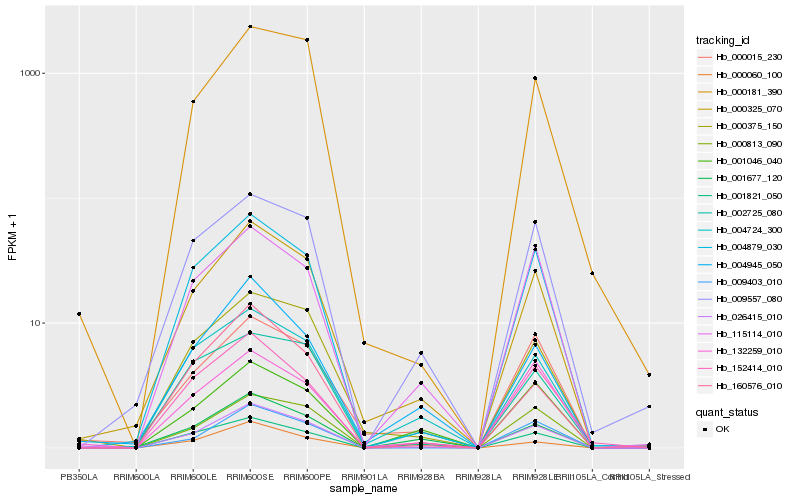
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026415_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000325_070 |
0.0730041986 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_004879_030 |
0.0777942055 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 4 |
Hb_009403_010 |
0.0966374199 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 5 |
Hb_000813_090 |
0.1021149453 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 6 |
Hb_115114_010 |
0.1126794096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_152414_010 |
0.113664683 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 8 |
Hb_009557_080 |
0.1173574397 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 9 |
Hb_000375_150 |
0.1177905316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000181_390 |
0.1194221256 |
- |
- |
unknown [Lotus japonicus] |
| 11 |
Hb_000060_100 |
0.1202619347 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004724_300 |
0.1213979937 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 13 |
Hb_001046_040 |
0.1283598212 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_001821_050 |
0.1294582773 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 15 |
Hb_001677_120 |
0.1316017534 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 16 |
Hb_004945_050 |
0.1329245017 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 17 |
Hb_000015_230 |
0.1362605312 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 18 |
Hb_132259_010 |
0.1368893573 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 19 |
Hb_160576_010 |
0.139235475 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002725_080 |
0.1398922683 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |