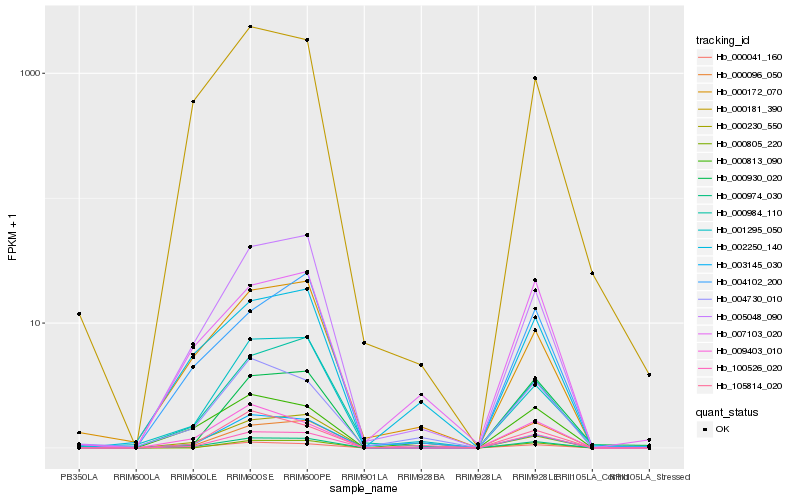
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000974_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105128463 isoform X1 [Populus euphratica] |
| 2 |
Hb_100526_020 |
0.0888614925 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 3 |
Hb_003145_030 |
0.1007087163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005048_090 |
0.1069249646 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 5 |
Hb_000096_050 |
0.1168276076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000181_390 |
0.1313694497 |
- |
- |
unknown [Lotus japonicus] |
| 7 |
Hb_009403_010 |
0.1326365 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 8 |
Hb_000984_110 |
0.1368285242 |
- |
- |
hypothetical protein JCGZ_15953 [Jatropha curcas] |
| 9 |
Hb_000230_550 |
0.1389810348 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001295_050 |
0.1442977293 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 11 |
Hb_000172_070 |
0.1475246726 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 12 |
Hb_000813_090 |
0.1509019188 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 13 |
Hb_004102_200 |
0.1534084694 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_000930_020 |
0.1538062241 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 15 |
Hb_000041_160 |
0.1561476242 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_000805_220 |
0.156750314 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 17 |
Hb_002250_140 |
0.157533788 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 18 |
Hb_004730_010 |
0.1595213189 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 19 |
Hb_105814_020 |
0.1609219093 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 20 |
Hb_007103_020 |
0.1619075764 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |