| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
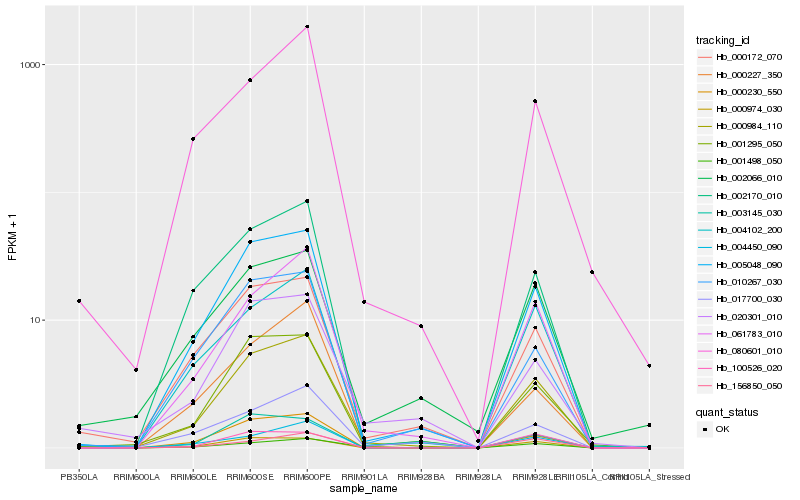
Hb_000984_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_15953 [Jatropha curcas] |
| 2 |
Hb_005048_090 |
0.0834939511 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 3 |
Hb_061783_010 |
0.1077232794 |
- |
- |
PREDICTED: uncharacterized protein LOC105124983 isoform X4 [Populus euphratica] |
| 4 |
Hb_000230_550 |
0.1122549457 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001295_050 |
0.1123865064 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 6 |
Hb_000172_070 |
0.1186378483 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_004102_200 |
0.1262799174 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_002170_010 |
0.1339658562 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 9 |
Hb_000974_030 |
0.1368285242 |
- |
- |
PREDICTED: uncharacterized protein LOC105128463 isoform X1 [Populus euphratica] |
| 10 |
Hb_080601_010 |
0.1389696639 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 11 |
Hb_020301_010 |
0.1422789632 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_010267_030 |
0.1430279808 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_002066_010 |
0.1437381444 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004450_090 |
0.1461185262 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 15 |
Hb_001498_050 |
0.1465178877 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 16 |
Hb_003145_030 |
0.1495133202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_017700_030 |
0.1560463302 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 18 |
Hb_100526_020 |
0.1563297864 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 19 |
Hb_000227_350 |
0.1594973207 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 20 |
Hb_156850_050 |
0.1614058525 |
- |
- |
PREDICTED: uncharacterized protein LOC105635250 [Jatropha curcas] |