| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
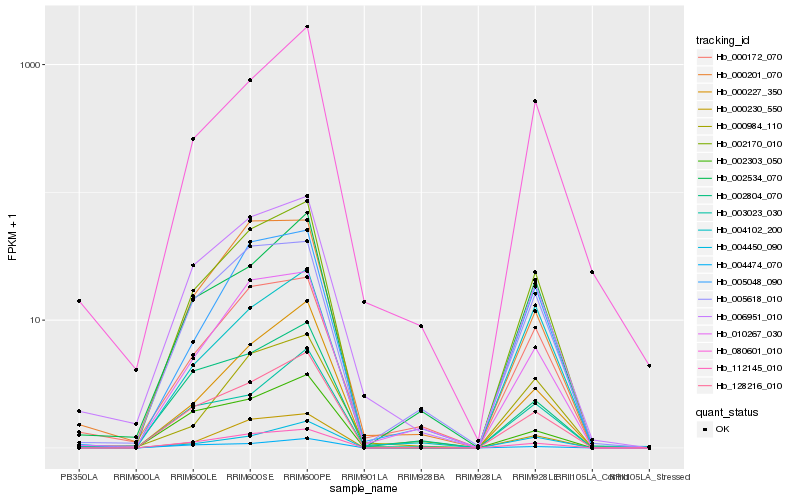
Hb_002170_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 2 |
Hb_128216_010 |
0.0672098346 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 3 |
Hb_112145_010 |
0.0723326009 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_005048_090 |
0.0958893194 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 5 |
Hb_010267_030 |
0.1005834417 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_006951_010 |
0.1067060038 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |
| 7 |
Hb_002534_070 |
0.1075525838 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 8 |
Hb_000230_550 |
0.1082273509 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005618_010 |
0.1116719045 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 10 |
Hb_000172_070 |
0.1118030849 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 11 |
Hb_000227_350 |
0.1136373273 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 12 |
Hb_080601_010 |
0.1157623679 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 13 |
Hb_004450_090 |
0.120056193 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 14 |
Hb_003023_030 |
0.1236210924 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 15 |
Hb_004102_200 |
0.1284493034 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_004474_070 |
0.1299141235 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP8, putative [Ricinus communis] |
| 17 |
Hb_000201_070 |
0.130692412 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 18 |
Hb_002303_050 |
0.1331151009 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor PTL-like [Populus euphratica] |
| 19 |
Hb_000984_110 |
0.1339658562 |
- |
- |
hypothetical protein JCGZ_15953 [Jatropha curcas] |
| 20 |
Hb_002804_070 |
0.1342064785 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |