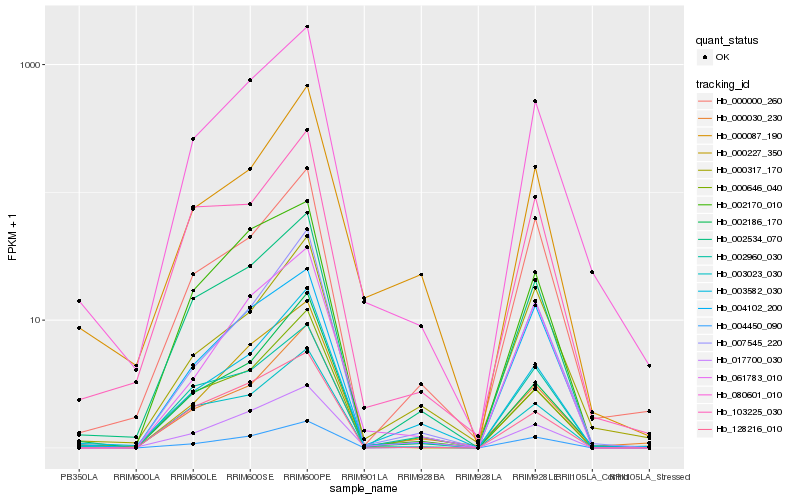
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080601_010 |
0.0 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 2 |
Hb_002534_070 |
0.0974645948 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 3 |
Hb_003023_030 |
0.1017790594 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_000030_230 |
0.1062527805 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 5 |
Hb_004450_090 |
0.1066249779 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 6 |
Hb_002186_170 |
0.1095062056 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 7 |
Hb_000000_260 |
0.1154248462 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 8 |
Hb_061783_010 |
0.1155135012 |
- |
- |
PREDICTED: uncharacterized protein LOC105124983 isoform X4 [Populus euphratica] |
| 9 |
Hb_002170_010 |
0.1157623679 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 10 |
Hb_003582_030 |
0.1157997023 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 11 |
Hb_004102_200 |
0.1182072279 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_000227_350 |
0.119320289 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 13 |
Hb_103225_030 |
0.1210190423 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000646_040 |
0.1244634027 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 15 |
Hb_007545_220 |
0.1251327179 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 16 |
Hb_128216_010 |
0.1286736357 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 17 |
Hb_017700_030 |
0.1295790583 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 18 |
Hb_000317_170 |
0.1298960093 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 19 |
Hb_002960_030 |
0.13142108 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 20 |
Hb_000087_190 |
0.1321449621 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |