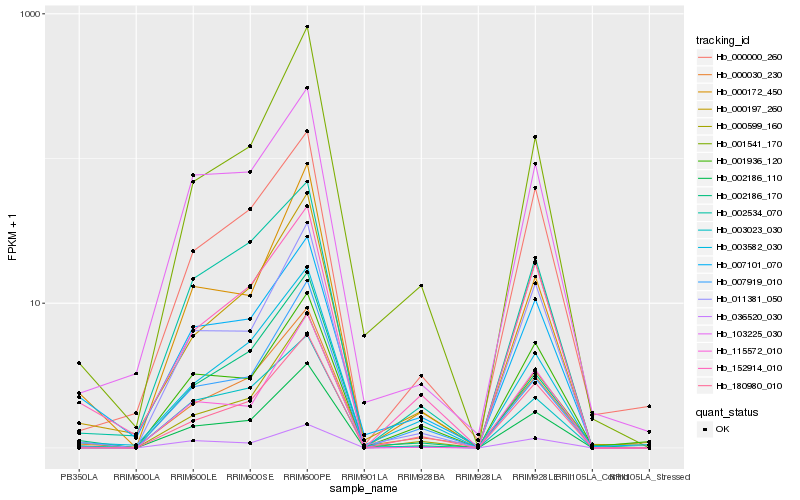
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002186_110 |
0.0 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 2 |
Hb_011381_050 |
0.0748544152 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 3 |
Hb_000197_260 |
0.0807343222 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 4 |
Hb_115572_010 |
0.0921926561 |
- |
- |
PREDICTED: beta-galactosidase 6 [Jatropha curcas] |
| 5 |
Hb_152914_010 |
0.0941135526 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 6 |
Hb_000172_450 |
0.0991266707 |
- |
- |
Uncharacterized protein TCM_001203 [Theobroma cacao] |
| 7 |
Hb_180980_010 |
0.0997911925 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 8 |
Hb_003582_030 |
0.1008289477 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 9 |
Hb_000030_230 |
0.103070666 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 10 |
Hb_003023_030 |
0.1034218007 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_103225_030 |
0.1043526747 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_001541_170 |
0.1054163001 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 13 |
Hb_001936_120 |
0.1081138842 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002534_070 |
0.1086939748 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 15 |
Hb_000000_260 |
0.1097873299 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 16 |
Hb_002186_170 |
0.1098104615 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 17 |
Hb_000599_160 |
0.1098687187 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 18 |
Hb_007101_070 |
0.110496535 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 19 |
Hb_007919_010 |
0.1106471948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_036520_030 |
0.1113490591 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |