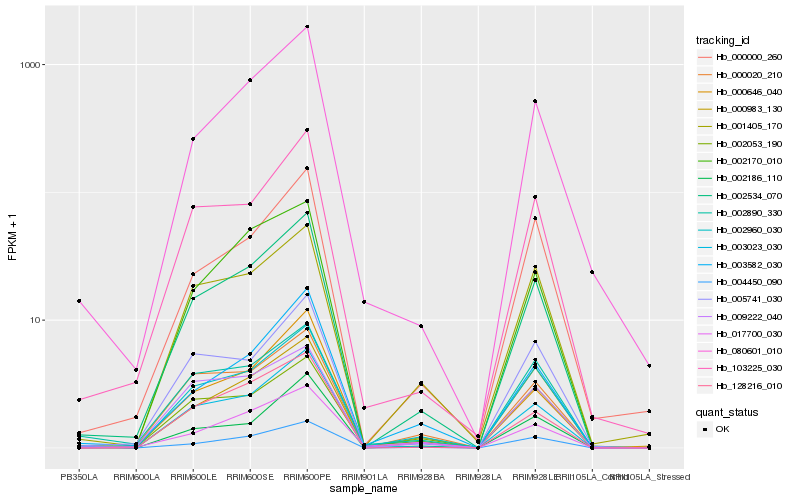
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002534_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 2 |
Hb_003023_030 |
0.0641694503 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 3 |
Hb_002960_030 |
0.0735091906 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 4 |
Hb_004450_090 |
0.0809660997 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 5 |
Hb_103225_030 |
0.0884560589 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000646_040 |
0.0926737052 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 7 |
Hb_080601_010 |
0.0974645948 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 8 |
Hb_128216_010 |
0.098688996 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 9 |
Hb_000020_210 |
0.0996677169 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 10 |
Hb_000000_260 |
0.1061499189 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 11 |
Hb_003582_030 |
0.1061584168 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 12 |
Hb_002170_010 |
0.1075525838 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 13 |
Hb_002186_110 |
0.1086939748 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 14 |
Hb_001405_170 |
0.1091121248 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 15 |
Hb_005741_030 |
0.1094835708 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 16 |
Hb_000983_130 |
0.109774541 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_009222_040 |
0.1098235561 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 18 |
Hb_002890_330 |
0.1151669528 |
- |
- |
caspase, putative [Ricinus communis] |
| 19 |
Hb_017700_030 |
0.1152749511 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 20 |
Hb_002053_190 |
0.1156098417 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |