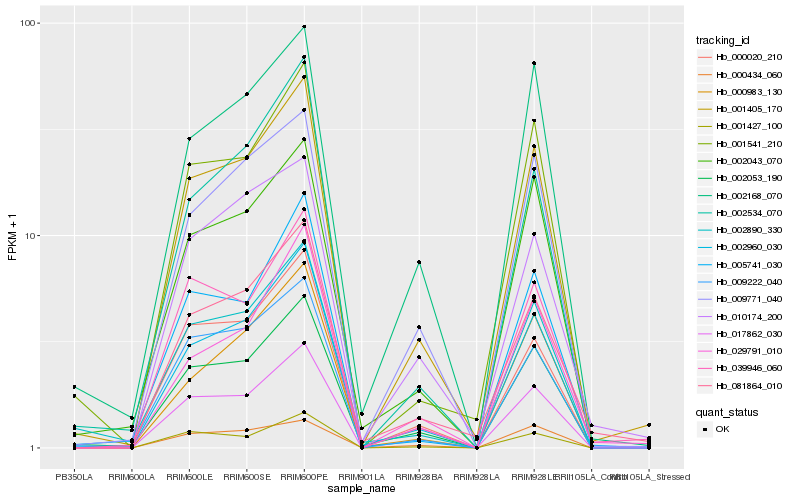
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_170 |
0.0 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 2 |
Hb_002053_190 |
0.0638507722 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002960_030 |
0.0658026899 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 4 |
Hb_081864_010 |
0.0834235682 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001541_210 |
0.0856002438 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 6 |
Hb_009771_040 |
0.0860964034 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 7 |
Hb_002168_070 |
0.0914745769 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002043_070 |
0.0921184549 |
- |
- |
PREDICTED: jacalin-related lectin 19 [Jatropha curcas] |
| 9 |
Hb_039946_060 |
0.0927073378 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 10 |
Hb_000020_210 |
0.093241315 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 11 |
Hb_009222_040 |
0.0952253964 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 12 |
Hb_017862_030 |
0.0963182978 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 13 |
Hb_002890_330 |
0.0984073005 |
- |
- |
caspase, putative [Ricinus communis] |
| 14 |
Hb_001427_100 |
0.10104101 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 15 |
Hb_005741_030 |
0.1084598438 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 16 |
Hb_010174_200 |
0.1090472391 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002534_070 |
0.1091121248 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 18 |
Hb_029791_010 |
0.110012789 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 19 |
Hb_000983_130 |
0.1119477028 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_000434_060 |
0.1130241893 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |