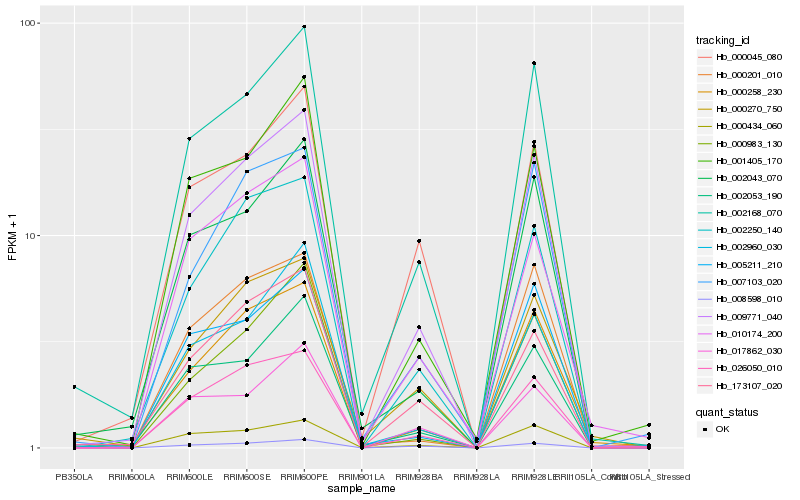
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009771_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 2 |
Hb_002168_070 |
0.059634713 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_026050_010 |
0.071768771 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 4 |
Hb_002250_140 |
0.073174485 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 5 |
Hb_000258_230 |
0.0789783635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000434_060 |
0.0845139805 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 7 |
Hb_173107_020 |
0.085242892 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 8 |
Hb_001405_170 |
0.0860964034 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 9 |
Hb_002043_070 |
0.0866454759 |
- |
- |
PREDICTED: jacalin-related lectin 19 [Jatropha curcas] |
| 10 |
Hb_000045_080 |
0.0940705448 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_002053_190 |
0.0942776285 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007103_020 |
0.0988852499 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 13 |
Hb_002960_030 |
0.1019184207 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 14 |
Hb_000270_750 |
0.1047545797 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 15 |
Hb_010174_200 |
0.1063434786 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008598_010 |
0.1068106886 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 17 |
Hb_000201_010 |
0.1089334296 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_005211_210 |
0.1089733167 |
- |
- |
lysine and histidine specific transporter family protein [Populus trichocarpa] |
| 19 |
Hb_017862_030 |
0.1093578412 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 20 |
Hb_000983_130 |
0.1096919453 |
- |
- |
transcription factor, putative [Ricinus communis] |