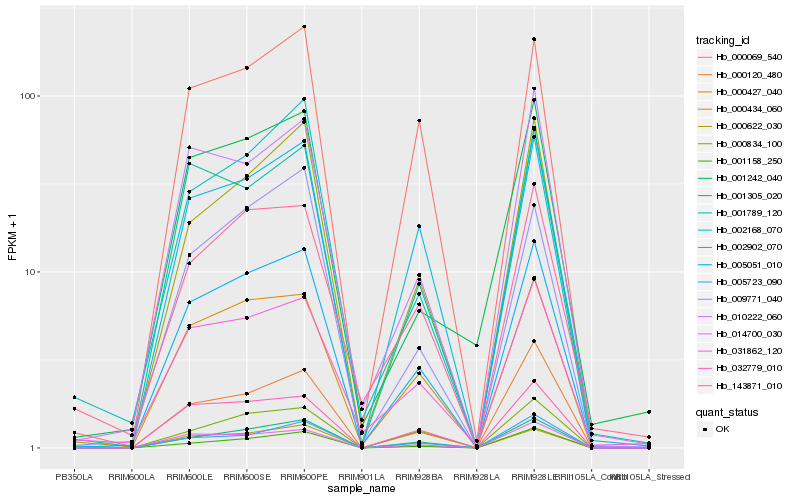
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005051_010 |
0.0 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001305_020 |
0.0623047738 |
- |
- |
PREDICTED: uncharacterized protein LOC102623247 [Citrus sinensis] |
| 3 |
Hb_000427_040 |
0.0624868654 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 4 |
Hb_014700_030 |
0.0838755417 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 5 |
Hb_032779_010 |
0.0859403465 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000622_030 |
0.0869654125 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 7 |
Hb_000434_060 |
0.0897160142 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 8 |
Hb_002902_070 |
0.0904358899 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 9 |
Hb_031862_120 |
0.0990339365 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 10 |
Hb_000069_540 |
0.0996602302 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 11 |
Hb_010222_060 |
0.1023348854 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_001158_250 |
0.103310802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001789_120 |
0.1052557251 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_001242_040 |
0.1064117765 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 15 |
Hb_005723_090 |
0.1076801438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_143871_010 |
0.108794598 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_002168_070 |
0.1106368286 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000120_480 |
0.1109526691 |
- |
- |
- |
| 19 |
Hb_000834_100 |
0.1114518598 |
- |
- |
PREDICTED: uncharacterized protein LOC105178355 [Sesamum indicum] |
| 20 |
Hb_009771_040 |
0.1121812743 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |