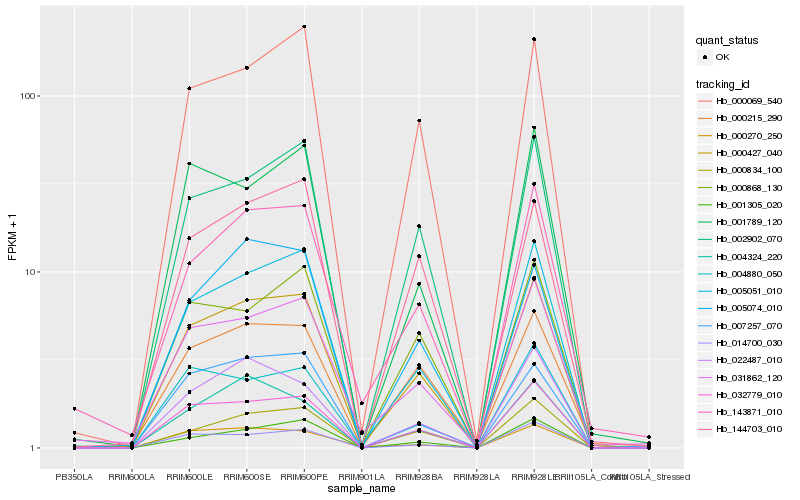
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000427_040 |
0.0 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 2 |
Hb_032779_010 |
0.0516202858 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_005051_010 |
0.0624868654 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_002902_070 |
0.0794552603 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 5 |
Hb_014700_030 |
0.0908579417 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 6 |
Hb_001305_020 |
0.0920778135 |
- |
- |
PREDICTED: uncharacterized protein LOC102623247 [Citrus sinensis] |
| 7 |
Hb_000069_540 |
0.0925534622 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 8 |
Hb_031862_120 |
0.1010652883 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 9 |
Hb_000215_290 |
0.1016065152 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000834_100 |
0.1018058861 |
- |
- |
PREDICTED: uncharacterized protein LOC105178355 [Sesamum indicum] |
| 11 |
Hb_022487_010 |
0.1042150195 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 12 |
Hb_004880_050 |
0.107043929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000868_130 |
0.1072015959 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 14 |
Hb_143871_010 |
0.1074135769 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 15 |
Hb_007257_070 |
0.1087452698 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 16 |
Hb_000270_250 |
0.112958395 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s07900g [Populus trichocarpa] |
| 17 |
Hb_001789_120 |
0.1140087801 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004324_220 |
0.114178661 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 19 |
Hb_144703_010 |
0.1142994227 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 20 |
Hb_005074_010 |
0.1149162421 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |