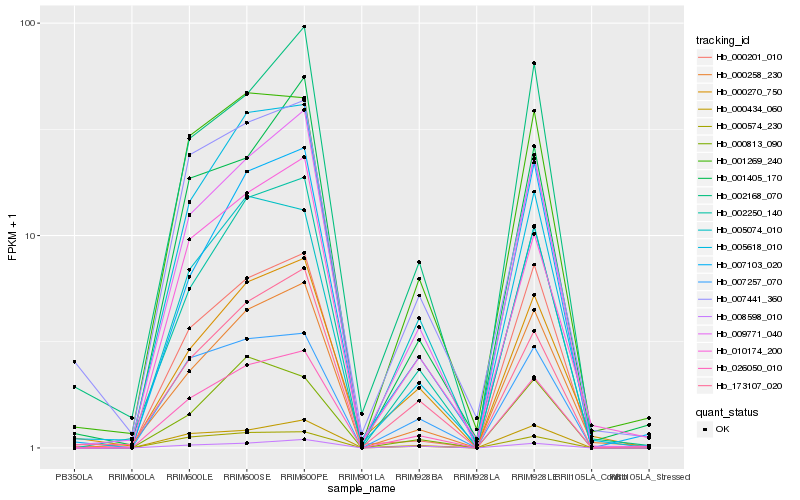
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026050_010 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_009771_040 |
0.071768771 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 3 |
Hb_002250_140 |
0.073310343 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 4 |
Hb_000574_230 |
0.0860052102 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EXS-like isoform X2 [Vitis vinifera] |
| 5 |
Hb_007257_070 |
0.0879439403 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 6 |
Hb_010174_200 |
0.0904221627 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000258_230 |
0.092712387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001269_240 |
0.0966913096 |
- |
- |
aquaporin NIP6.1 family protein [Populus trichocarpa] |
| 9 |
Hb_173107_020 |
0.1009323032 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 10 |
Hb_000434_060 |
0.1077188276 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 11 |
Hb_007103_020 |
0.1096270912 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 12 |
Hb_001405_170 |
0.1149315402 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 13 |
Hb_005074_010 |
0.1165549818 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 14 |
Hb_002168_070 |
0.118743438 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_008598_010 |
0.1219238674 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 16 |
Hb_007441_360 |
0.1223817496 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 17 |
Hb_000270_750 |
0.124012998 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 18 |
Hb_005618_010 |
0.1240286976 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 19 |
Hb_000201_010 |
0.1242730683 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_000813_090 |
0.1260299085 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |