| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
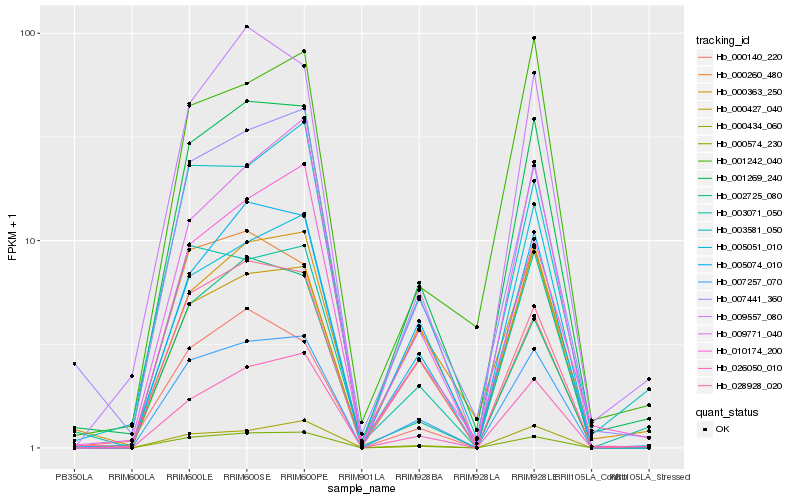
Hb_001269_240 |
0.0 |
- |
- |
aquaporin NIP6.1 family protein [Populus trichocarpa] |
| 2 |
Hb_007257_070 |
0.062442442 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 3 |
Hb_000574_230 |
0.0723734816 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EXS-like isoform X2 [Vitis vinifera] |
| 4 |
Hb_026050_010 |
0.0966913096 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 5 |
Hb_009557_080 |
0.0980634125 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 6 |
Hb_007441_360 |
0.1014527207 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 7 |
Hb_005074_010 |
0.107792152 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 8 |
Hb_028928_020 |
0.1079269845 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 9 |
Hb_000140_220 |
0.1108486084 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 10 |
Hb_002725_080 |
0.1113347276 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_003071_050 |
0.116236679 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 12 |
Hb_000434_060 |
0.1171934272 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 13 |
Hb_003581_050 |
0.1195926778 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 14 |
Hb_000427_040 |
0.1247932545 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 15 |
Hb_000363_250 |
0.1250587221 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 16 |
Hb_005051_010 |
0.1269282553 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_010174_200 |
0.1283576553 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009771_040 |
0.1298309582 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 19 |
Hb_001242_040 |
0.130814581 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 20 |
Hb_000260_480 |
0.1311487189 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |