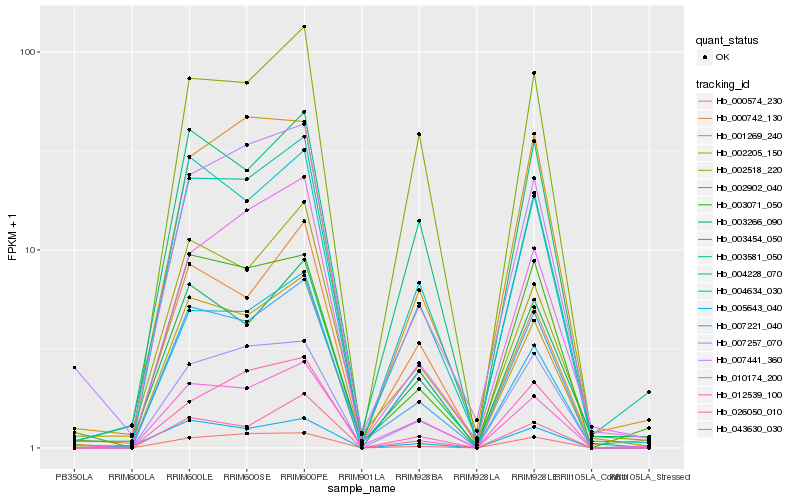
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003581_050 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 2 |
Hb_003266_090 |
0.0979451797 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 3 |
Hb_007221_040 |
0.1023586821 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 4 |
Hb_002518_220 |
0.1035032966 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 5 |
Hb_007441_360 |
0.1044464257 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 6 |
Hb_010174_200 |
0.1074442198 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007257_070 |
0.1089523184 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 8 |
Hb_004228_070 |
0.1102177572 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 9 |
Hb_002205_150 |
0.1111764917 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 10 |
Hb_000742_130 |
0.1161615774 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 11 |
Hb_012539_100 |
0.1191662095 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |
| 12 |
Hb_001269_240 |
0.1195926778 |
- |
- |
aquaporin NIP6.1 family protein [Populus trichocarpa] |
| 13 |
Hb_003071_050 |
0.1223179263 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 14 |
Hb_000574_230 |
0.1246448521 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EXS-like isoform X2 [Vitis vinifera] |
| 15 |
Hb_004634_030 |
0.124932174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002902_040 |
0.1256753996 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 17 |
Hb_005643_040 |
0.1280467872 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_003454_050 |
0.1281319965 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 19 |
Hb_026050_010 |
0.1288915441 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 20 |
Hb_043630_030 |
0.1289145126 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |