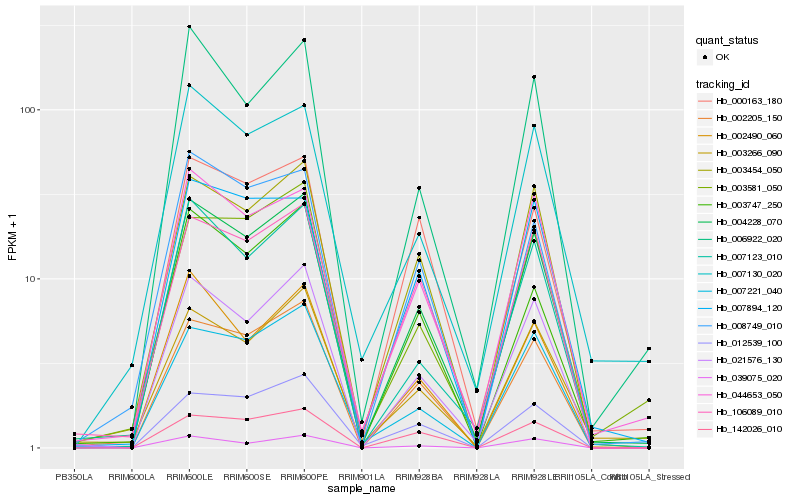
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004228_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 2 |
Hb_021576_130 |
0.0619604528 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002205_150 |
0.072368675 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 4 |
Hb_003454_050 |
0.0750874004 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 5 |
Hb_007123_010 |
0.0866779987 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 6 |
Hb_044653_050 |
0.0873837544 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006922_020 |
0.0884456869 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 8 |
Hb_007221_040 |
0.0891418326 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 9 |
Hb_142026_010 |
0.0912302046 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Jatropha curcas] |
| 10 |
Hb_002490_060 |
0.0912788543 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 11 |
Hb_039075_020 |
0.0948098576 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 12 |
Hb_106089_010 |
0.0975819585 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 13 |
Hb_008749_010 |
0.1007898175 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 14 |
Hb_003747_250 |
0.1016820815 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 15 |
Hb_003266_090 |
0.1019967531 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 16 |
Hb_000163_180 |
0.10529573 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 17 |
Hb_007894_120 |
0.1064122437 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_007130_020 |
0.1077724777 |
- |
- |
- |
| 19 |
Hb_012539_100 |
0.1087307406 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK11 [Jatropha curcas] |
| 20 |
Hb_003581_050 |
0.1102177572 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |