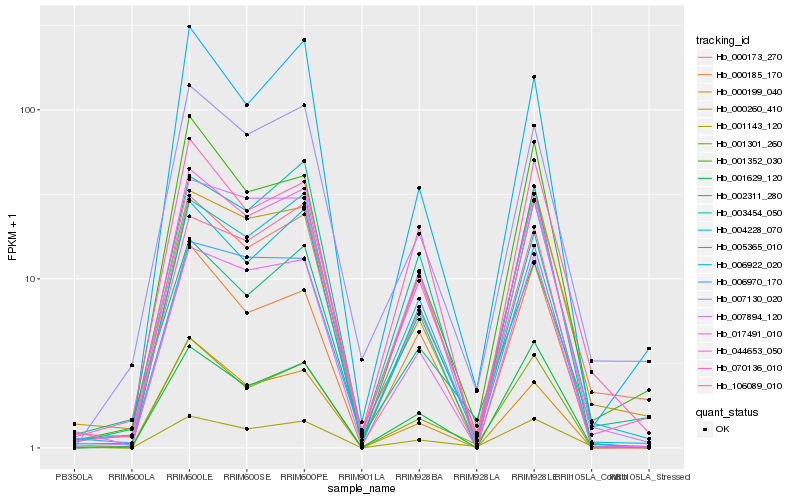
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044653_050 |
0.0 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007130_020 |
0.0756212877 |
- |
- |
- |
| 3 |
Hb_007894_120 |
0.080727792 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_017491_010 |
0.0811184664 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004228_070 |
0.0873837544 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 6 |
Hb_005365_010 |
0.0914306289 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 7 |
Hb_001301_260 |
0.0937125689 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000260_410 |
0.0964141843 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 9 |
Hb_000173_270 |
0.0968782961 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003454_050 |
0.0971764963 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 11 |
Hb_002311_280 |
0.0980384115 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 12 |
Hb_001629_120 |
0.098315647 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 13 |
Hb_006922_020 |
0.0997553481 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 14 |
Hb_000199_040 |
0.1067252153 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_106089_010 |
0.1070742711 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 16 |
Hb_001352_030 |
0.1075538007 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 17 |
Hb_001143_120 |
0.1076064372 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 18 |
Hb_070136_010 |
0.1078191179 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_000185_170 |
0.108087175 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_006970_170 |
0.1092590243 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |