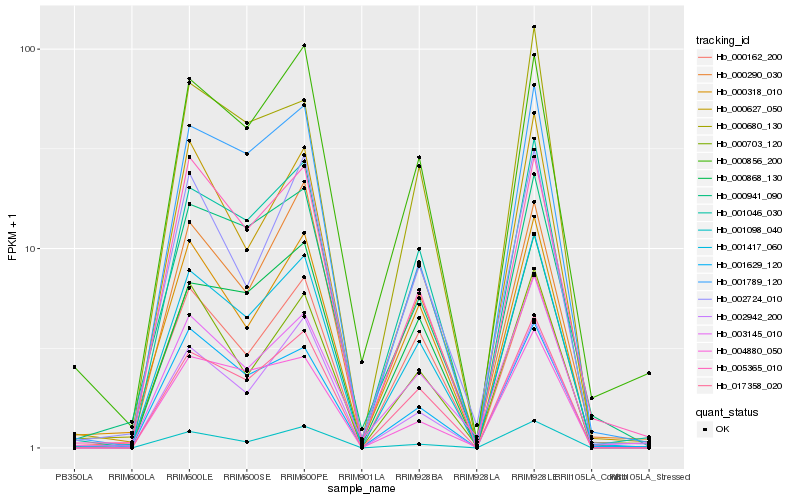
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001417_060 |
0.0 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 2 |
Hb_001046_030 |
0.0535238966 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_017358_020 |
0.0556887776 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 4 |
Hb_000868_130 |
0.0844028769 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 5 |
Hb_001789_120 |
0.0853112455 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000627_050 |
0.0854258746 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 7 |
Hb_000941_090 |
0.0879815452 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 8 |
Hb_001629_120 |
0.0894720005 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 9 |
Hb_000318_010 |
0.0911181475 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 10 |
Hb_005365_010 |
0.0911246744 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 11 |
Hb_000162_200 |
0.0937261386 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001098_040 |
0.0941786221 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 13 |
Hb_000703_120 |
0.0956784564 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000856_200 |
0.0965962785 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 15 |
Hb_002942_200 |
0.0968788191 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH137 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003145_010 |
0.0969639854 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 17 |
Hb_002724_010 |
0.0972594088 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 18 |
Hb_000680_130 |
0.0977786523 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 19 |
Hb_004880_050 |
0.10081793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000290_030 |
0.1011252346 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |