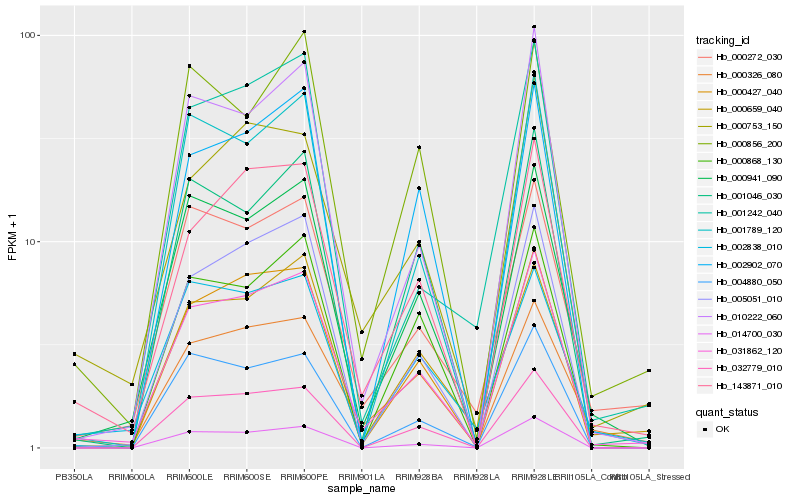
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031862_120 |
0.0 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 2 |
Hb_143871_010 |
0.0787053042 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 3 |
Hb_000941_090 |
0.0884415529 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 4 |
Hb_005051_010 |
0.0990339365 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000753_150 |
0.0999596635 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 6 |
Hb_000427_040 |
0.1010652883 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 7 |
Hb_000272_030 |
0.1040793219 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010222_060 |
0.1060225071 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000326_080 |
0.1061984706 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_032779_010 |
0.1087482734 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001789_120 |
0.1116959263 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000659_040 |
0.1126554734 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_014700_030 |
0.1147764668 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 14 |
Hb_001046_030 |
0.1177165193 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_004880_050 |
0.1197115033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000868_130 |
0.121776321 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 17 |
Hb_002902_070 |
0.122220197 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 18 |
Hb_001242_040 |
0.1226392099 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 19 |
Hb_000856_200 |
0.1226825214 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 20 |
Hb_002838_010 |
0.1240108305 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |