| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
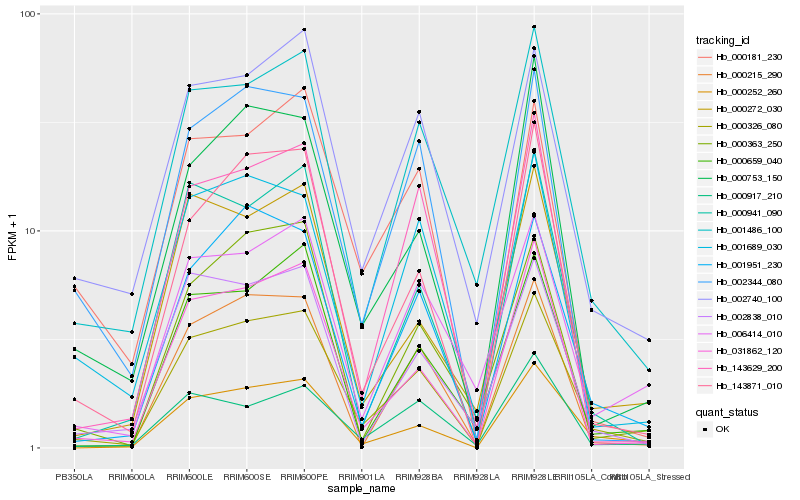
Hb_000326_080 |
0.0 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_001486_100 |
0.0994900043 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 3 |
Hb_000252_260 |
0.1047456735 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 4 |
Hb_031862_120 |
0.1061984706 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 5 |
Hb_001951_230 |
0.1066594032 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 6 |
Hb_143871_010 |
0.1134386688 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 7 |
Hb_002740_100 |
0.1171012098 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_000215_290 |
0.1232432027 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002344_080 |
0.1257316117 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 10 |
Hb_143629_200 |
0.1262679143 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 11 |
Hb_000272_030 |
0.1265579372 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000659_040 |
0.1266802313 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002838_010 |
0.1274692926 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 14 |
Hb_000941_090 |
0.1280424465 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 15 |
Hb_001689_030 |
0.128201442 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 16 |
Hb_006414_010 |
0.1292689968 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 17 |
Hb_000753_150 |
0.1306909682 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 18 |
Hb_000917_210 |
0.1365212651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000363_250 |
0.1368907402 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 20 |
Hb_000181_230 |
0.1377884638 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |