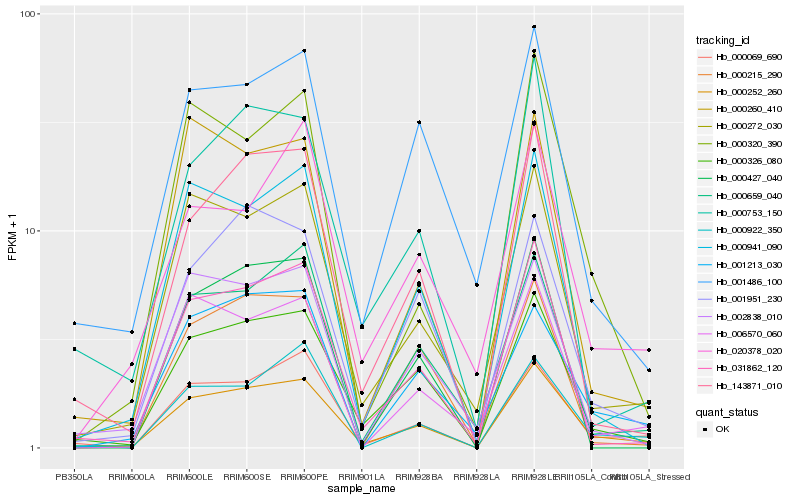
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_260 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 2 |
Hb_000326_080 |
0.1047456735 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000069_690 |
0.117130335 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 4 |
Hb_000272_030 |
0.1234258469 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 5 |
Hb_031862_120 |
0.1299041342 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 6 |
Hb_001951_230 |
0.135600637 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 7 |
Hb_000659_040 |
0.1359188984 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000922_350 |
0.1376657291 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 9 |
Hb_000941_090 |
0.1381561021 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 10 |
Hb_143871_010 |
0.1395749802 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 11 |
Hb_000320_390 |
0.1458718397 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 12 |
Hb_006570_060 |
0.1465319761 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 13 |
Hb_001213_030 |
0.1469100129 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 14 |
Hb_001486_100 |
0.15491594 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 15 |
Hb_000260_410 |
0.1558850475 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 16 |
Hb_000215_290 |
0.1562433617 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_020378_020 |
0.157668258 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000427_040 |
0.1588688266 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 19 |
Hb_000753_150 |
0.160633095 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 20 |
Hb_002838_010 |
0.1610123309 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |