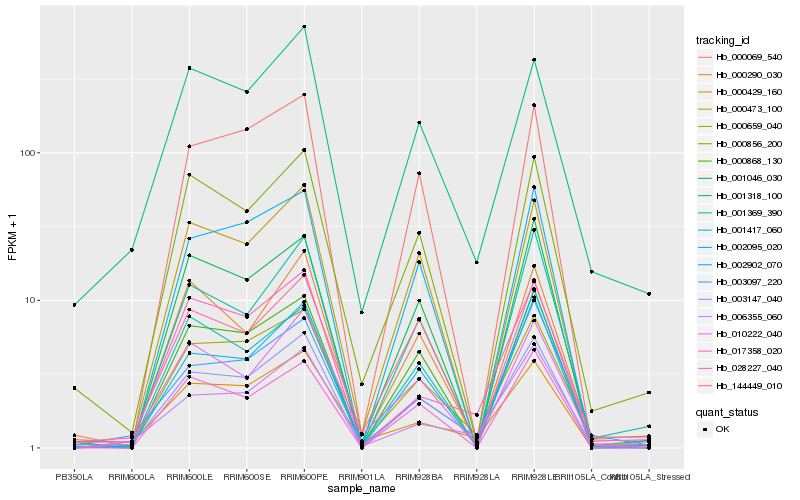
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003147_040 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 2 |
Hb_000659_040 |
0.0925410364 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002095_020 |
0.1038524844 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001369_390 |
0.1093919368 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 5 |
Hb_002902_070 |
0.1241155078 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 6 |
Hb_000473_100 |
0.1244466687 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 7 |
Hb_000868_130 |
0.124914217 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 8 |
Hb_000069_540 |
0.1254088752 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 9 |
Hb_000290_030 |
0.126050885 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 10 |
Hb_028227_040 |
0.1262782288 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 11 |
Hb_006355_060 |
0.1268393433 |
- |
- |
PREDICTED: transcription factor PIF1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001046_030 |
0.1276735589 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001318_100 |
0.1287759492 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 14 |
Hb_000856_200 |
0.1292160146 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 15 |
Hb_000429_160 |
0.130244453 |
- |
- |
PREDICTED: uncharacterized protein LOC105645177 [Jatropha curcas] |
| 16 |
Hb_017358_020 |
0.1317981369 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 17 |
Hb_003097_220 |
0.1322607576 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 18 |
Hb_001417_060 |
0.1326588286 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 19 |
Hb_010222_040 |
0.1344333921 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |
| 20 |
Hb_144449_010 |
0.136449891 |
- |
- |
Potassium transporter, putative [Ricinus communis] |