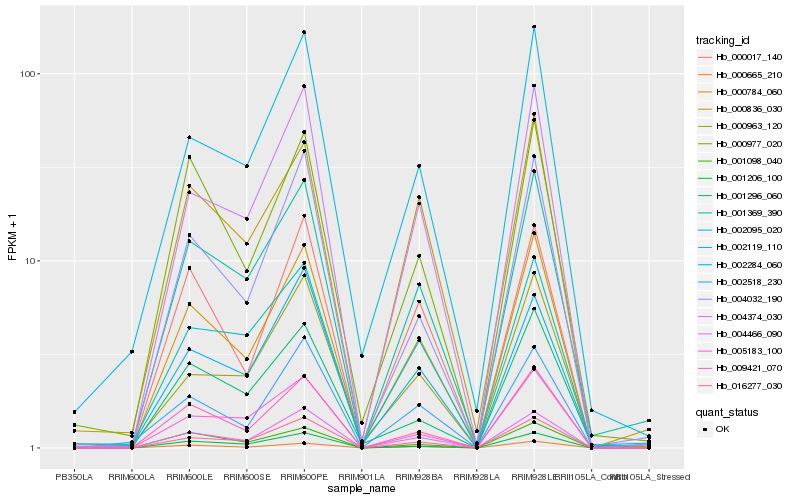
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000784_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000665_210 |
0.0638942136 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 3 |
Hb_004466_090 |
0.0658270551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009421_070 |
0.0711542683 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 5 |
Hb_001369_390 |
0.0715643472 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 6 |
Hb_004374_030 |
0.0719901275 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 7 |
Hb_016277_030 |
0.0788091213 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 8 |
Hb_004032_190 |
0.0894731667 |
- |
- |
PREDICTED: probable cellulose synthase A catalytic subunit 3 [UDP-forming] isoform X1 [Jatropha curcas] |
| 9 |
Hb_000836_030 |
0.0929926104 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 10 |
Hb_002095_020 |
0.0947660758 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002119_110 |
0.0972462851 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 12 |
Hb_000017_140 |
0.0979092418 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 13 |
Hb_000977_020 |
0.1008277744 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 14 |
Hb_001206_100 |
0.1040973799 |
- |
- |
hypothetical protein PRUPE_ppa021915mg, partial [Prunus persica] |
| 15 |
Hb_002518_230 |
0.1101170531 |
- |
- |
- |
| 16 |
Hb_001296_060 |
0.1104221659 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_001098_040 |
0.1104896301 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 18 |
Hb_005183_100 |
0.1112259789 |
- |
- |
hypothetical protein CICLE_v10002898mg [Citrus clementina] |
| 19 |
Hb_002284_060 |
0.1113637216 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 20 |
Hb_000963_120 |
0.1132569787 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |