| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
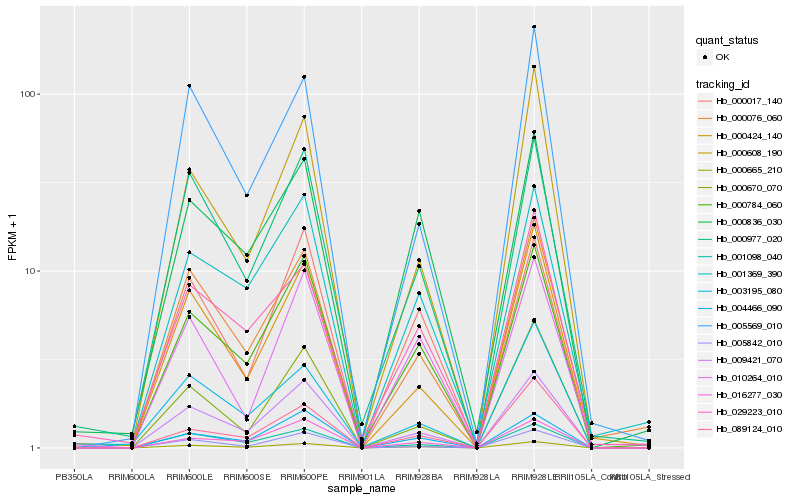
Hb_000665_210 |
0.0 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 2 |
Hb_009421_070 |
0.0598198097 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 3 |
Hb_000784_060 |
0.0638942136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001098_040 |
0.0922394309 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 5 |
Hb_000076_060 |
0.0922908513 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 6 |
Hb_000424_140 |
0.0930100211 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000977_020 |
0.0956844192 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 8 |
Hb_004466_090 |
0.1005487238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000017_140 |
0.1038674613 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 10 |
Hb_000670_070 |
0.1055849634 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 11 |
Hb_000836_030 |
0.106068014 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 12 |
Hb_010264_010 |
0.1064187574 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 13 |
Hb_000608_190 |
0.1085768883 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 14 |
Hb_001369_390 |
0.1088782684 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 15 |
Hb_005569_010 |
0.1104571019 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 16 |
Hb_089124_010 |
0.1110192981 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 17 |
Hb_005842_010 |
0.1111622647 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 18 |
Hb_016277_030 |
0.1121752337 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 19 |
Hb_029223_010 |
0.1139585551 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 20 |
Hb_003195_080 |
0.1142234794 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |