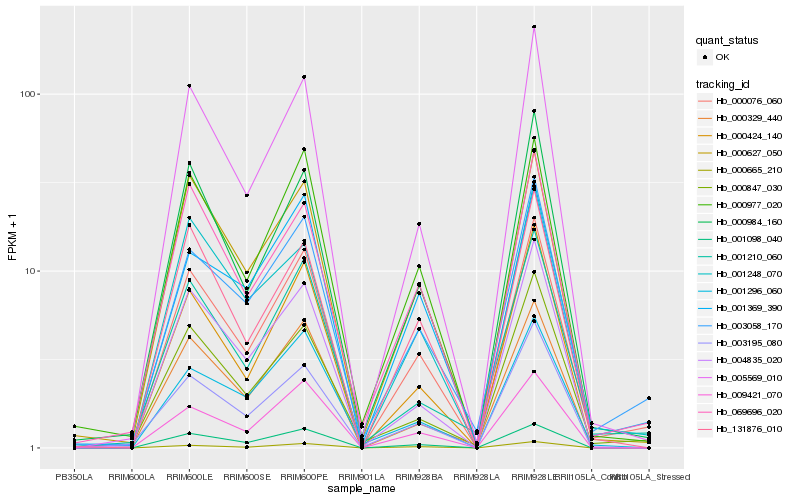
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000076_060 |
0.0 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 2 |
Hb_003058_170 |
0.0795964348 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005569_010 |
0.0825442943 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 4 |
Hb_001098_040 |
0.0862950771 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 5 |
Hb_000665_210 |
0.0922908513 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 6 |
Hb_000977_020 |
0.0928390997 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 7 |
Hb_009421_070 |
0.0932309521 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 8 |
Hb_000424_140 |
0.093686521 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000984_160 |
0.0942022158 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000627_050 |
0.0986013446 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 11 |
Hb_131876_010 |
0.0991567657 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_069696_020 |
0.0993609056 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001210_060 |
0.101447942 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 14 |
Hb_004835_020 |
0.1030548893 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 15 |
Hb_000329_440 |
0.1055513237 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 16 |
Hb_001248_070 |
0.1073492457 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 17 |
Hb_001296_060 |
0.1103102209 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 18 |
Hb_000847_030 |
0.11066039 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 19 |
Hb_003195_080 |
0.111838607 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 20 |
Hb_001369_390 |
0.1131224876 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |