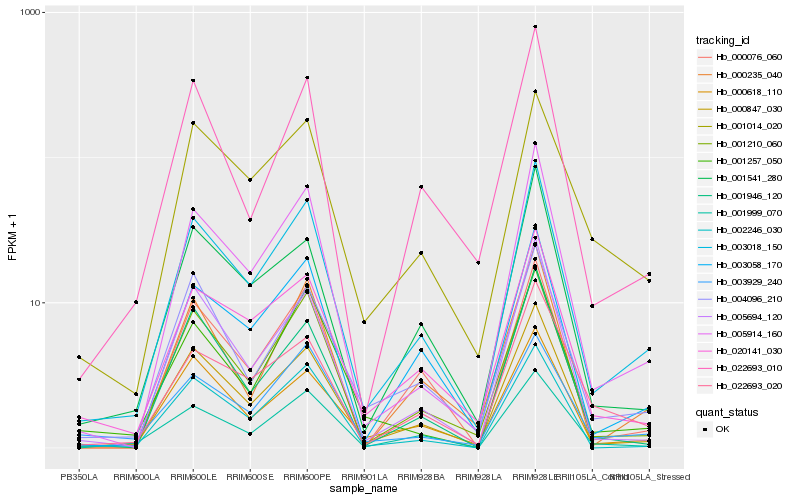
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003018_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 2 |
Hb_000847_030 |
0.0707106213 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 3 |
Hb_005914_160 |
0.084830344 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001210_060 |
0.0856652834 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 5 |
Hb_003058_170 |
0.0860654958 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 6 |
Hb_020141_030 |
0.1025731249 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_000618_110 |
0.1064823765 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 8 |
Hb_005694_120 |
0.10670986 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 9 |
Hb_001999_070 |
0.1075216761 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 10 |
Hb_002246_030 |
0.1115778532 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_001541_280 |
0.1134184613 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 12 |
Hb_000076_060 |
0.1168094102 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 13 |
Hb_004096_210 |
0.1184495002 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 14 |
Hb_022693_010 |
0.1200054971 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 15 |
Hb_003929_240 |
0.1214563224 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 16 |
Hb_022693_020 |
0.1218624868 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 17 |
Hb_001257_050 |
0.1256916822 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein YLS9 [Jatropha curcas] |
| 18 |
Hb_000235_040 |
0.1264234859 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X1 [Populus euphratica] |
| 19 |
Hb_001014_020 |
0.1267239533 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 20 |
Hb_001946_120 |
0.1280180142 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |