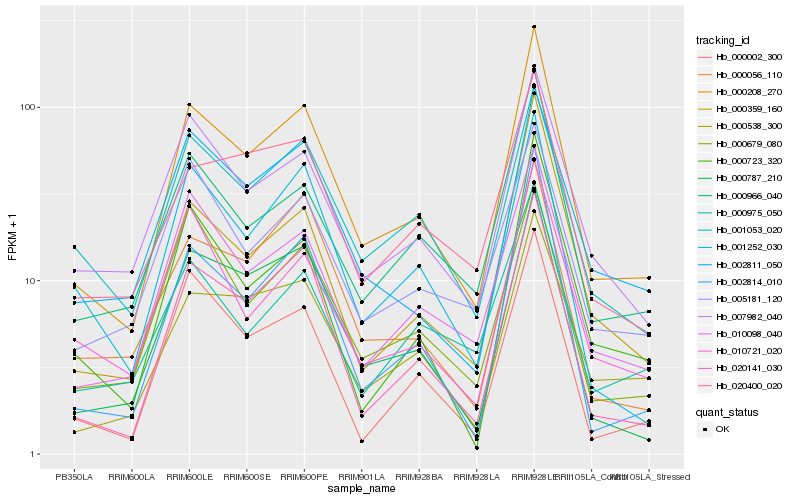
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000208_270 |
0.0 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 2 |
Hb_020141_030 |
0.0850743713 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_010098_040 |
0.1021097838 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 4 |
Hb_000787_210 |
0.1033964992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000538_300 |
0.1076601649 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000966_040 |
0.108973507 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 7 |
Hb_002814_010 |
0.1103795909 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 8 |
Hb_007982_040 |
0.1133004882 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000359_160 |
0.1141119888 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 10 |
Hb_001252_030 |
0.1196082975 |
- |
- |
Serine/threonine-protein kinase Nek8, putative [Ricinus communis] |
| 11 |
Hb_000002_300 |
0.1196357516 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_000723_320 |
0.1234244916 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 13 |
Hb_002811_050 |
0.1237742383 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 14 |
Hb_001053_020 |
0.1245969605 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 15 |
Hb_000056_110 |
0.1269159978 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 16 |
Hb_005181_120 |
0.1273208236 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000679_080 |
0.1275940629 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_020400_020 |
0.1289622212 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 19 |
Hb_010721_020 |
0.1294414854 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000975_050 |
0.1315992415 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |