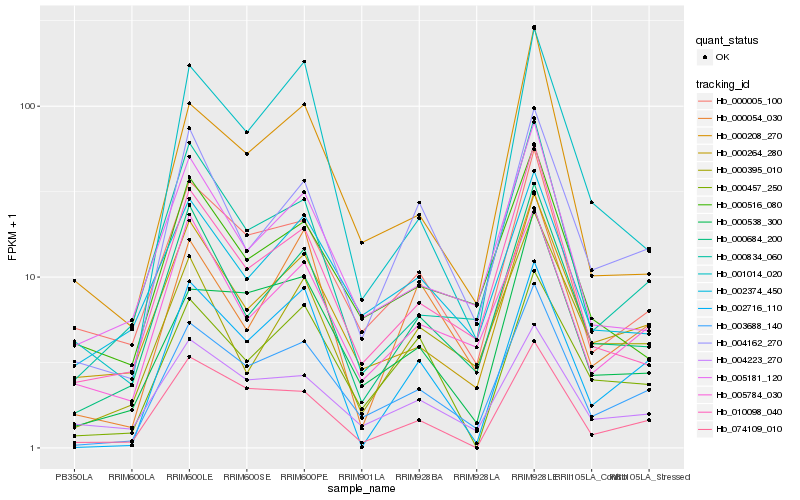
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 2 |
Hb_000538_300 |
0.1190038973 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000264_280 |
0.1270593244 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 4 |
Hb_000684_200 |
0.1274236458 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 5 |
Hb_000005_100 |
0.1280746817 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004162_270 |
0.1287418418 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_010098_040 |
0.1332410539 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 8 |
Hb_074109_010 |
0.1359183638 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 9 |
Hb_000834_060 |
0.1399203567 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_005784_030 |
0.1432905174 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004223_270 |
0.1436345479 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000208_270 |
0.1444617218 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 13 |
Hb_005181_120 |
0.1465959843 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000457_250 |
0.1494131862 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000054_030 |
0.150547382 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 16 |
Hb_002374_450 |
0.1516028642 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002716_110 |
0.1521832174 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 18 |
Hb_000516_080 |
0.1541087301 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 19 |
Hb_000395_010 |
0.1551068712 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 20 |
Hb_001014_020 |
0.1564689353 |
- |
- |
gene GA family protein [Populus trichocarpa] |