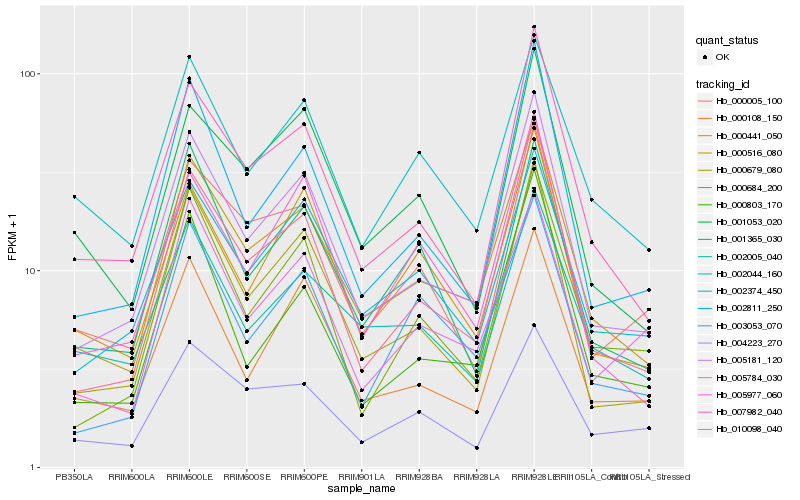
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000516_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 2 |
Hb_005181_120 |
0.0643060227 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_010098_040 |
0.0786247267 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 4 |
Hb_007982_040 |
0.0897609497 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000679_080 |
0.0984187254 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002374_450 |
0.1039743895 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002811_250 |
0.1077816615 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002044_160 |
0.1084679697 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 9 |
Hb_003053_070 |
0.1088399687 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000005_100 |
0.1103305599 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000441_050 |
0.1127065834 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 12 |
Hb_002005_040 |
0.1127459776 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000108_150 |
0.1138006352 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 14 |
Hb_001053_020 |
0.116569895 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 15 |
Hb_004223_270 |
0.1167934788 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005784_030 |
0.1176131646 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000684_200 |
0.1176700759 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 18 |
Hb_001365_030 |
0.1177436826 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 19 |
Hb_005977_060 |
0.1183300562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000803_170 |
0.1188084021 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |