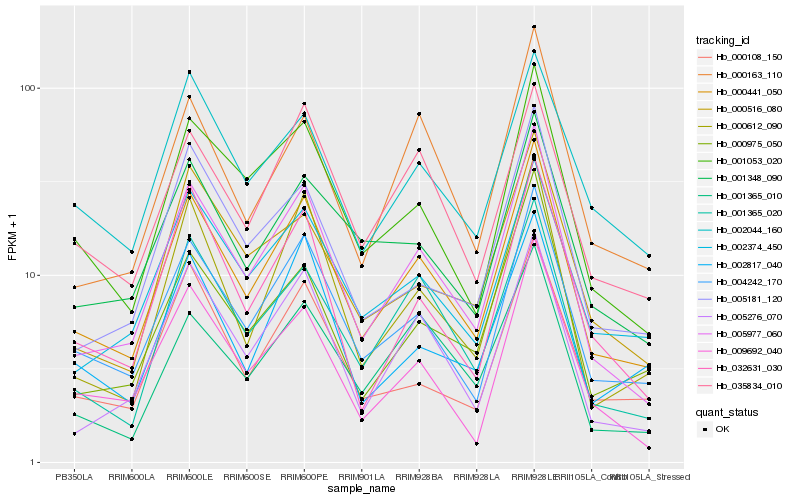
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000441_050 |
0.0 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 2 |
Hb_005977_060 |
0.0563589724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004242_170 |
0.0573848704 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 4 |
Hb_001053_020 |
0.0890094496 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 5 |
Hb_001365_020 |
0.0938603411 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 6 |
Hb_002374_450 |
0.0959499811 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005276_070 |
0.0975430229 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 8 |
Hb_000612_090 |
0.1003669692 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 9 |
Hb_001348_090 |
0.1013153276 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 10 |
Hb_009692_040 |
0.1021425977 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000108_150 |
0.1031784657 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 12 |
Hb_032631_030 |
0.1051254812 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 13 |
Hb_035834_010 |
0.1056735883 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_002044_160 |
0.1068427906 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 15 |
Hb_000163_110 |
0.1070106416 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 16 |
Hb_005181_120 |
0.110598708 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_001365_010 |
0.1119036597 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 18 |
Hb_000975_050 |
0.1120193079 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000516_080 |
0.1127065834 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 20 |
Hb_002817_040 |
0.1145527794 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |