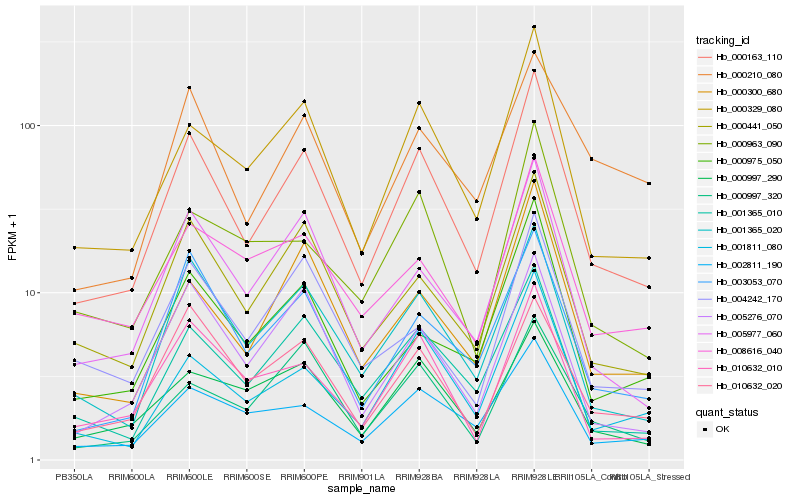
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000163_110 |
0.0 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 2 |
Hb_000329_080 |
0.094427362 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001365_020 |
0.1006418092 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 4 |
Hb_001365_010 |
0.1059668492 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 5 |
Hb_000441_050 |
0.1070106416 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 6 |
Hb_005977_060 |
0.1147846869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000300_680 |
0.1176492138 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001811_080 |
0.1180975194 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 9 |
Hb_003053_070 |
0.1181877222 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002811_190 |
0.1185891796 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 11 |
Hb_010632_010 |
0.123902379 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_005276_070 |
0.1259196556 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 13 |
Hb_000975_050 |
0.1288004491 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_010632_020 |
0.1374991204 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 15 |
Hb_000997_290 |
0.1383081973 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 16 |
Hb_000997_320 |
0.1401474733 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 17 |
Hb_000963_090 |
0.1402482667 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_008616_040 |
0.1451135188 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004242_170 |
0.1452059589 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 20 |
Hb_000210_080 |
0.145861398 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |