| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
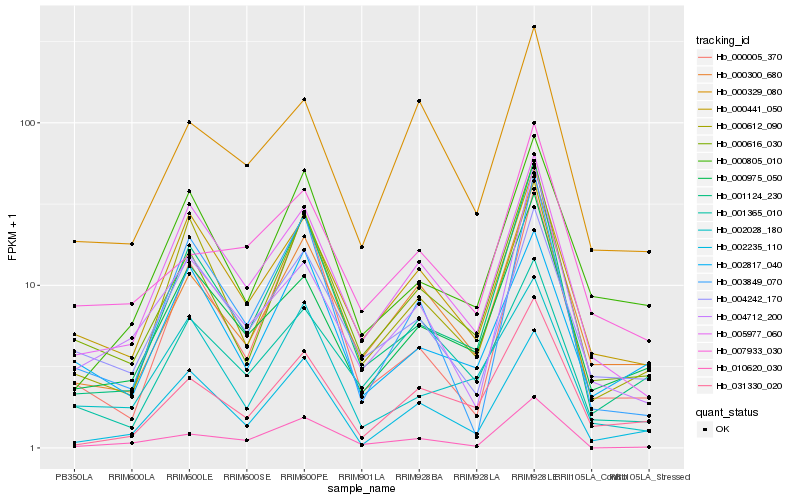
Hb_000616_030 |
0.0 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 2 |
Hb_000300_680 |
0.0872239224 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001124_230 |
0.1044313321 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000441_050 |
0.1277489163 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 5 |
Hb_000612_090 |
0.1308508595 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 6 |
Hb_005977_060 |
0.1336887139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000975_050 |
0.1352013761 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_031330_020 |
0.1378300101 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 9 |
Hb_002028_180 |
0.1396547882 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_004242_170 |
0.1397301928 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 11 |
Hb_002235_110 |
0.1430302258 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 12 |
Hb_002817_040 |
0.1449268978 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 13 |
Hb_000805_010 |
0.1453239928 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 14 |
Hb_000005_370 |
0.14653949 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 15 |
Hb_010620_030 |
0.15441929 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 16 |
Hb_004712_200 |
0.1547495696 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 17 |
Hb_001365_010 |
0.1558301862 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 18 |
Hb_007933_030 |
0.1560709048 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 19 |
Hb_003849_070 |
0.1561090546 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 20 |
Hb_000329_080 |
0.1571780085 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |