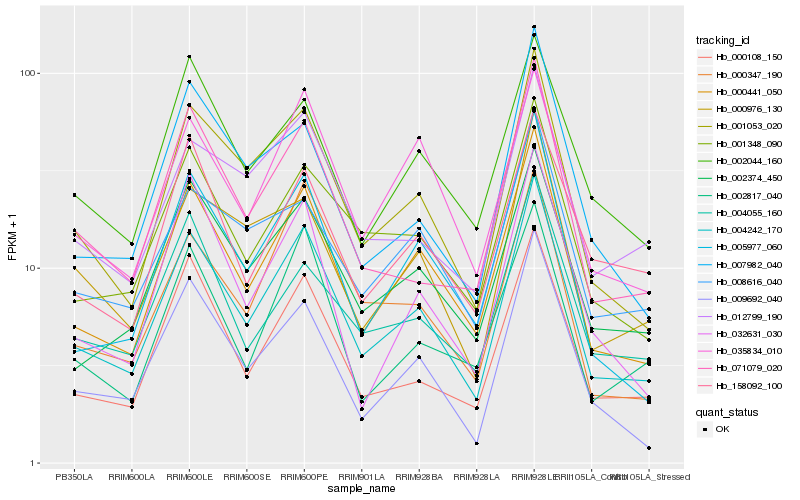
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004242_170 |
0.0 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 2 |
Hb_000441_050 |
0.0573848704 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 3 |
Hb_001053_020 |
0.0736484433 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 4 |
Hb_000108_150 |
0.0907634169 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 5 |
Hb_005977_060 |
0.0931507962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001348_090 |
0.0941016195 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009692_040 |
0.0957709237 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002374_450 |
0.0969100036 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_071079_020 |
0.0999489287 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 10 |
Hb_012799_190 |
0.1062473701 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 11 |
Hb_002817_040 |
0.110809314 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 12 |
Hb_000976_130 |
0.1116723376 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 13 |
Hb_035834_010 |
0.1120075882 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000347_190 |
0.1126860555 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 15 |
Hb_032631_030 |
0.1137598184 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 16 |
Hb_002044_160 |
0.1142614534 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 17 |
Hb_008616_040 |
0.1150278714 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004055_160 |
0.1155694682 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_158092_100 |
0.1172414207 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 20 |
Hb_007982_040 |
0.117448483 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |