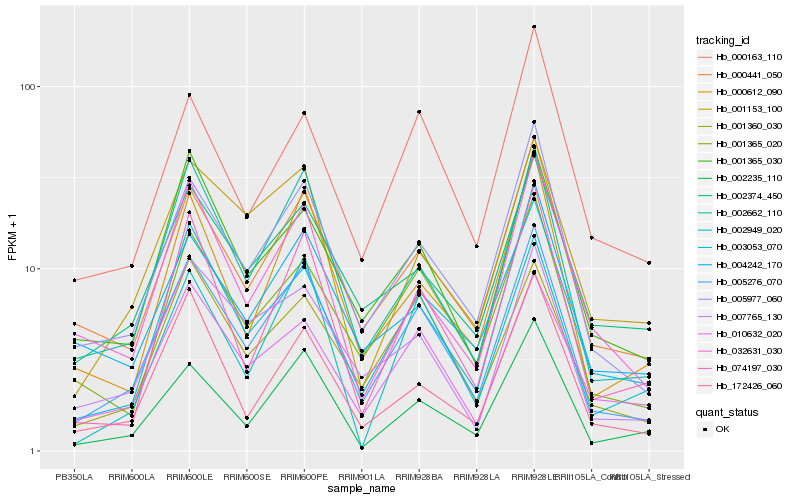
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005276_070 |
0.0 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 2 |
Hb_005977_060 |
0.07867125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002662_110 |
0.0900020893 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_000441_050 |
0.0975430229 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 5 |
Hb_001365_020 |
0.1053856469 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 6 |
Hb_003053_070 |
0.1109715025 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_010632_020 |
0.1129046284 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 8 |
Hb_001360_030 |
0.1141747089 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 9 |
Hb_007765_130 |
0.1157820505 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002235_110 |
0.1158035113 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 11 |
Hb_000612_090 |
0.1162652439 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 12 |
Hb_001365_030 |
0.1182517548 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 13 |
Hb_074197_030 |
0.120520221 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_002374_450 |
0.1215703502 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000163_110 |
0.1259196556 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 16 |
Hb_032631_030 |
0.1272607504 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 17 |
Hb_004242_170 |
0.1272669474 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 18 |
Hb_001153_100 |
0.1282417874 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 19 |
Hb_002949_020 |
0.1302413497 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 20 |
Hb_172426_060 |
0.1308014166 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |