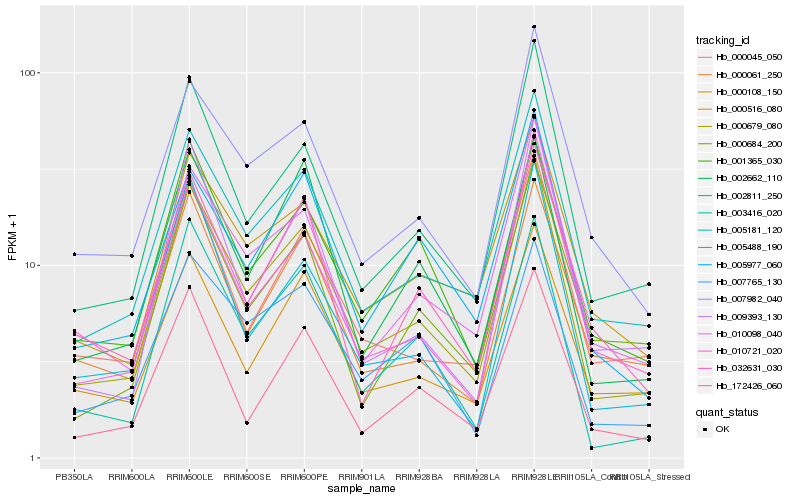
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_080 |
0.0 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005181_120 |
0.0650679655 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002811_250 |
0.0752839945 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_010098_040 |
0.0826275551 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 5 |
Hb_003416_020 |
0.0936988104 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 6 |
Hb_000516_080 |
0.0984187254 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 7 |
Hb_007982_040 |
0.0984384118 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000045_050 |
0.0995192854 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002662_110 |
0.1027527663 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_000108_150 |
0.1027708374 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 11 |
Hb_172426_060 |
0.103771401 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 12 |
Hb_001365_030 |
0.1053880323 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 13 |
Hb_000061_250 |
0.1062482406 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 14 |
Hb_007765_130 |
0.1089847113 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 15 |
Hb_009393_130 |
0.1092593783 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 16 |
Hb_005488_190 |
0.109305562 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 17 |
Hb_010721_020 |
0.1112441712 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_005977_060 |
0.1121007284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_032631_030 |
0.1133399482 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 20 |
Hb_000684_200 |
0.1141299611 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |